+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1dyq | ||||||
|---|---|---|---|---|---|---|---|
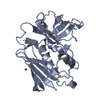
| Title | STAPHYLOCOCCAL ENTEROTOXIN A MUTANT VACCINE | ||||||
 Components Components | Enterotoxin type A | ||||||
 Keywords Keywords | ENTEROTOXIN / STAPHYLOCOCCAL ENTEROTOXIN TYPE A / VACCINE | ||||||
| Function / homology |  Function and homology information Function and homology informationMHC class II protein binding / toxin activity / extracellular region / metal ion binding Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.5 Å MOLECULAR REPLACEMENT / Resolution: 1.5 Å | ||||||
 Authors Authors | Krupka, H.I. / Segelke, B.W. / Rupp, B. | ||||||
 Citation Citation |  Journal: Protein Sci. / Year: 2002 Journal: Protein Sci. / Year: 2002Title: Structural Basis for Abrogated Binding between Staphylococcal Enterotoxin a Superantigen Vaccine and Mhc-II? Authors: Krupka, H.I. / Segelke, B.W. / Ulrich, R. / Ringhofer, S. / Knapp, M. / Rupp, B. | ||||||
| History |
| ||||||
| Remark 700 | SHEET DETERMINATION METHOD: THERE IS A BIFURCATED SHEET IN THIS STRUCTURE. THIS SHEET REPRESENTED ... SHEET DETERMINATION METHOD: THERE IS A BIFURCATED SHEET IN THIS STRUCTURE. THIS SHEET REPRESENTED BY TWO SHEETS WHICH HAS TWO IDENTICAL STRANDS. SHEETS A AND A1 REPRESENT ONE BIFURCATED SHEET. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1dyq.cif.gz 1dyq.cif.gz | 72.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1dyq.ent.gz pdb1dyq.ent.gz | 53.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1dyq.json.gz 1dyq.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1dyq_validation.pdf.gz 1dyq_validation.pdf.gz | 427.2 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1dyq_full_validation.pdf.gz 1dyq_full_validation.pdf.gz | 432.3 KB | Display | |
| Data in XML |  1dyq_validation.xml.gz 1dyq_validation.xml.gz | 17 KB | Display | |
| Data in CIF |  1dyq_validation.cif.gz 1dyq_validation.cif.gz | 26.8 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/dy/1dyq https://data.pdbj.org/pub/pdb/validation_reports/dy/1dyq ftp://data.pdbj.org/pub/pdb/validation_reports/dy/1dyq ftp://data.pdbj.org/pub/pdb/validation_reports/dy/1dyq | HTTPS FTP |
-Related structure data
| Related structure data |  1esfS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Details | BIOLOGICAL UNIT: MONOMER |
- Components
Components
| #1: Protein | Mass: 27255.547 Da / Num. of mol.: 1 / Mutation: YES Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|---|
| #2: Chemical | ChemComp-ZN / |
| #3: Chemical | ChemComp-SO4 / |
| #4: Water | ChemComp-HOH / |
| Has protein modification | Y |
| Sequence details | VACCINE MUTANT L48R, Y92A, D70R |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.5 Å3/Da / Density % sol: 51 % | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 277 K / Method: vapor diffusion, hanging drop / pH: 7.8 Details: HANGING DROP OVER 1 ML WELL, 4 + 4 UL DROPS, WELL SOLUTION 10% PEG6K, 5% MPD IN 0.1 M HEPES PH 7.8, TEMPERATURE 4 DEG, 3-4 WEEKS. | ||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 4 ℃ / Method: vapor diffusion, hanging drop / pH: 7.8 | ||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 120 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ALS ALS  / Beamline: 5.0.2 / Wavelength: 1.1 / Beamline: 5.0.2 / Wavelength: 1.1 |
| Detector | Type: ADSC QUANTUM 4 / Detector: CCD / Date: Sep 15, 1999 / Details: DOUBLE FOCUSSING |
| Radiation | Monochromator: SI / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.1 Å / Relative weight: 1 |
| Reflection | Resolution: 1.5→43.19 Å / Num. obs: 35972 / % possible obs: 82.8 % / Redundancy: 4.5 % / Biso Wilson estimate: 17.5 Å2 / Rsym value: 0.061 / Net I/σ(I): 7 |
| Reflection shell | Resolution: 1.5→1.54 Å / Redundancy: 2.8 % / Mean I/σ(I) obs: 4.4 / Rsym value: 0.135 / % possible all: 46.7 |
| Reflection | *PLUS Num. measured all: 158285 / Rmerge(I) obs: 0.062 |
| Reflection shell | *PLUS % possible obs: 37.2 % / Num. unique obs: 1143 / Num. measured obs: 2752 / Rmerge(I) obs: 0.135 / Mean I/σ(I) obs: 3.4 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1ESF Resolution: 1.5→43.187 Å / SU B: 1.726 / SU ML: 0.063 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.097 / ESU R Free: 0.101 Details: RESIDUES GLU 9, ASP 60 AND GLU 191 ARE IN WEAK DENSITY AND SIDE CHAIN ATOMS WERE NOT REFINED.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 20 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.5→43.187 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name: REFMAC / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Rfactor obs: 0.191 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller













 PDBj
PDBj






