+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1dpt | ||||||
|---|---|---|---|---|---|---|---|
| Title | D-DOPACHROME TAUTOMERASE | ||||||
 Components Components | D-DOPACHROME TAUTOMERASE | ||||||
 Keywords Keywords | CYTOKINE / GROWTH FACTOR / TAUTOMERASE | ||||||
| Function / homology |  Function and homology information Function and homology informationD-dopachrome decarboxylase / D-dopachrome decarboxylase activity / dopachrome isomerase activity / melanin biosynthetic process / phenylpyruvate tautomerase activity / cytokine receptor binding / negative regulation of macrophage chemotaxis / positive regulation of inflammatory response / positive regulation of tumor necrosis factor production / positive regulation of ERK1 and ERK2 cascade ...D-dopachrome decarboxylase / D-dopachrome decarboxylase activity / dopachrome isomerase activity / melanin biosynthetic process / phenylpyruvate tautomerase activity / cytokine receptor binding / negative regulation of macrophage chemotaxis / positive regulation of inflammatory response / positive regulation of tumor necrosis factor production / positive regulation of ERK1 and ERK2 cascade / extracellular space / extracellular exosome / cytoplasm Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.54 Å MOLECULAR REPLACEMENT / Resolution: 1.54 Å | ||||||
 Authors Authors | Sugimoto, H. / Taniguchi, M. / Nakagawa, A. / Tanaka, I. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 1999 Journal: Biochemistry / Year: 1999Title: Crystal structure of human D-dopachrome tautomerase, a homologue of macrophage migration inhibitory factor, at 1.54 A resolution. Authors: Sugimoto, H. / Taniguchi, M. / Nakagawa, A. / Tanaka, I. / Suzuki, M. / Nishihira, J. #1:  Journal: Biochem.Biophys.Res.Commun. / Year: 1998 Journal: Biochem.Biophys.Res.Commun. / Year: 1998Title: Molecular Cloning of Human D-Dopachrome Tautomerase Cdna: N-Terminal Proline is Essential for Enzyme Activation Authors: Nishihira, J. / Fujinaga, M. / Kuriyama, T. / Suzuki, M. / Sugimoto, H. / Nakagawa, A. / Tanaka, I. / Sakai, M. #2:  Journal: J.Struct.Biol. / Year: 1997 Journal: J.Struct.Biol. / Year: 1997Title: Crystallization and Preliminary X-Ray Analysis of Human D-Dopachrome Tautomerase Authors: Sugimoto, H. / Taniguchi, M. / Nakagawa, A. / Tanaka, I. / Suzuki, M. / Nishihira, J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1dpt.cif.gz 1dpt.cif.gz | 80.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1dpt.ent.gz pdb1dpt.ent.gz | 61.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1dpt.json.gz 1dpt.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1dpt_validation.pdf.gz 1dpt_validation.pdf.gz | 420.1 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1dpt_full_validation.pdf.gz 1dpt_full_validation.pdf.gz | 420.1 KB | Display | |
| Data in XML |  1dpt_validation.xml.gz 1dpt_validation.xml.gz | 16.3 KB | Display | |
| Data in CIF |  1dpt_validation.cif.gz 1dpt_validation.cif.gz | 23.7 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/dp/1dpt https://data.pdbj.org/pub/pdb/validation_reports/dp/1dpt ftp://data.pdbj.org/pub/pdb/validation_reports/dp/1dpt ftp://data.pdbj.org/pub/pdb/validation_reports/dp/1dpt | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 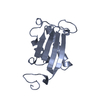
| ||||||||||||
| 2 | 
| ||||||||||||
| 3 | 
| ||||||||||||
| Unit cell |
| ||||||||||||
| Noncrystallographic symmetry (NCS) | NCS oper:
|
- Components
Components
| #1: Protein | Mass: 12593.526 Da / Num. of mol.: 3 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Cell line: BL21 / Organ: LIVER / Plasmid: PET3 / Production host: Homo sapiens (human) / Cell line: BL21 / Organ: LIVER / Plasmid: PET3 / Production host:  #2: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.21 Å3/Da / Density % sol: 45 % Description: DATA WERE COLLECTED USING THE WEISSENBERG METHOD. | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | pH: 5.7 / Details: pH 5.7 | ||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 18 ℃ / pH: 7.8 / Method: vapor diffusion, hanging drop | ||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 283 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Photon Factory Photon Factory  / Beamline: BL-6B / Wavelength: 1 / Beamline: BL-6B / Wavelength: 1 |
| Detector | Type: RIGAKU / Detector: IMAGE PLATE / Date: Nov 1, 1996 / Details: MIRROR |
| Radiation | Monochromator: SI(111) / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 1.54→20 Å / Num. obs: 43724 / % possible obs: 90.9 % / Observed criterion σ(I): 6 / Redundancy: 2.4 % / Biso Wilson estimate: 13.8 Å2 / Rmerge(I) obs: 0.036 / Net I/σ(I): 15.4 |
| Reflection shell | Resolution: 1.54→1.62 Å / Redundancy: 1.9 % / Rmerge(I) obs: 0.276 / Mean I/σ(I) obs: 2.4 / % possible all: 81.9 |
| Reflection | *PLUS Num. measured all: 104959 |
| Reflection shell | *PLUS % possible obs: 81.9 % |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: HUMAN MACROPHAGE MIGRATION INHIBITORY FACTOR Resolution: 1.54→20 Å / Rfactor Rfree error: 0.005 / Data cutoff high absF: 1000000 / Data cutoff low absF: 0.001 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 14.8 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.54→20 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints NCS | NCS model details: CONSTR | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.54→1.61 Å / Rfactor Rfree error: 0.022 / Total num. of bins used: 8
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Version: 3.851 / Classification: refinement X-PLOR / Version: 3.851 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | *PLUS Rfactor obs: 0.275 |
 Movie
Movie Controller
Controller













 PDBj
PDBj
