+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1ddy | ||||||
|---|---|---|---|---|---|---|---|
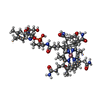
| Title | MOLECULAR RECOGNITION BY THE VITAMIN B12 RNA APTAMER | ||||||
 Components Components | VITAMIN B12 BINDING RNA | ||||||
 Keywords Keywords | RNA / TRIPLEX / VITAMIN B12 / APTAMER | ||||||
| Function / homology | COBALAMIN / METHYLAMINE / RNA / RNA (> 10) Function and homology information Function and homology information | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / MAD PHASING / Resolution: 3 Å SYNCHROTRON / MAD PHASING / Resolution: 3 Å | ||||||
 Authors Authors | Sussman, D. / Nix, J.C. / Wilson, C. | ||||||
 Citation Citation |  Journal: Nat.Struct.Biol. / Year: 2000 Journal: Nat.Struct.Biol. / Year: 2000Title: The structural basis for molecular recognition by the vitamin B 12 RNA aptamer. Authors: Sussman, D. / Nix, J.C. / Wilson, C. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1ddy.cif.gz 1ddy.cif.gz | 94.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1ddy.ent.gz pdb1ddy.ent.gz | 73.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1ddy.json.gz 1ddy.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1ddy_validation.pdf.gz 1ddy_validation.pdf.gz | 2.2 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1ddy_full_validation.pdf.gz 1ddy_full_validation.pdf.gz | 2.2 MB | Display | |
| Data in XML |  1ddy_validation.xml.gz 1ddy_validation.xml.gz | 8.9 KB | Display | |
| Data in CIF |  1ddy_validation.cif.gz 1ddy_validation.cif.gz | 11.1 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/dd/1ddy https://data.pdbj.org/pub/pdb/validation_reports/dd/1ddy ftp://data.pdbj.org/pub/pdb/validation_reports/dd/1ddy ftp://data.pdbj.org/pub/pdb/validation_reports/dd/1ddy | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||
| 2 | 
| ||||||||||
| 3 | 
| ||||||||||
| 4 | 
| ||||||||||
| Unit cell |
|
- Components
Components
| #1: RNA chain | Mass: 11280.819 Da / Num. of mol.: 4 / Source method: obtained synthetically Details: SEQUENCE RESULTS FROM IN VITRO SELECTION EXPERIMENTS #2: Chemical | ChemComp-B12 / #3: Chemical | ChemComp-NME / |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 4.13 Å3/Da / Density % sol: 70.25 % | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 289 K / Method: vapor diffusion, hanging drop / pH: 7.4 Details: LICL, ISOPROPANOL, MGCL2, pH 7.4, VAPOR DIFFUSION, HANGING DROP, temperature 289K | ||||||||||||||||||||||||||||||
| Components of the solutions |
| ||||||||||||||||||||||||||||||
| Crystal grow | *PLUS pH: 6.5 / Details: Sussman, D., (1999) Acta Cryst., D55, 326. | ||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ALS ALS  / Beamline: 5.0.1 / Wavelength: 1.6047,1.6057,1.5842 / Beamline: 5.0.1 / Wavelength: 1.6047,1.6057,1.5842 | ||||||||||||
| Detector | Type: ADSC QUANTUM 4 / Detector: CCD / Date: Sep 20, 1998 | ||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||
| Radiation wavelength |
| ||||||||||||
| Reflection | Resolution: 3→20 Å / Num. all: 40156 / Num. obs: 40156 / % possible obs: 92 % / Redundancy: 3.85 % / Rsym value: 0.061 / Net I/σ(I): 13.7 | ||||||||||||
| Reflection shell | Resolution: 3→3.05 Å / Redundancy: 2 % / Mean I/σ(I) obs: 4.469 / Rsym value: 0.195 / % possible all: 80 | ||||||||||||
| Reflection | *PLUS Highest resolution: 3 Å |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure: MAD PHASING / Resolution: 3→12 Å / Rfactor Rfree error: 0.01 / Data cutoff high rms absF: 3943390.88 / Cross valid method: THROUGHOUT / σ(F): 0 / σ(I): 0 Details: FOR CONSISTENCY, SUGARS PUCKERS WERE RESTRAINED TO THE A-RNA C3'-ENDO CONFORMATION. THIS MIGHT CONTRIBUTE TO THE HIGH RMSD FROM IDEAL VALUES OF IMPROPER ANGLES
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 26.5 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 3→12 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 3→3.18 Å / Rfactor Rfree error: 0.03 / Total num. of bins used: 6
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name: CNS / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Highest resolution: 3 Å / Lowest resolution: 12 Å / σ(F): 0 / % reflection Rfree: 10 % / Rfactor obs: 0.207 / Rfactor Rfree: 0.248 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | *PLUS Rfactor Rfree: 0.322 / % reflection Rfree: 5.9 % / Rfactor Rwork: 0.283 |
 Movie
Movie Controller
Controller








 PDBj
PDBj