[English] 日本語
 Yorodumi
Yorodumi- PDB-1cxr: AUTOMATED 2D NOESY ASSIGNMENT AND STRUCTURE CALCULATION OF CRAMBI... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1cxr | ||||||
|---|---|---|---|---|---|---|---|
| Title | AUTOMATED 2D NOESY ASSIGNMENT AND STRUCTURE CALCULATION OF CRAMBIN(S22/I25) WITH SELF-CORRECTING DISTANCE GEOMETRY BASED NOAH/DIAMOD PROGRAMS | ||||||
 Components Components | CRAMBIN | ||||||
 Keywords Keywords | PLANT PROTEIN / CRAMBIN / CRAMBE ABYSSINICA / PLANT SEED PROTEIN FEB. 20 / 1997 | ||||||
| Function / homology |  Function and homology information Function and homology information | ||||||
| Biological species |  Crambe hispanica subsp. abyssinica (Abyssinian crambe) Crambe hispanica subsp. abyssinica (Abyssinian crambe) | ||||||
| Method | SOLUTION NMR / SELF-CORRECTING DISTANCE GEOMETRY | ||||||
 Authors Authors | Xu, Y. / Wu, J. / Gorenstein, D. / Braun, W. | ||||||
 Citation Citation |  Journal: J.Magn.Reson. / Year: 1999 Journal: J.Magn.Reson. / Year: 1999Title: Automated 2D NOESY assignment and structure calculation of Crambin(S22/I25) with the self-correcting distance geometry based NOAH/DIAMOD programs. Authors: Xu, Y. / Wu, J. / Gorenstein, D. / Braun, W. #1:  Journal: J.Mol.Biol. / Year: 1995 Journal: J.Mol.Biol. / Year: 1995Title: Automated Assignment of Simulated, Experimental Noesy Spectra of Proteins by Feedback Filtering and Self-Correcting Distance Geometry Authors: Mumenthaler, C. / Braun, W. #2:  Journal: J.Biomol.NMR / Year: 1997 Journal: J.Biomol.NMR / Year: 1997Title: Automated Combined Assignment of Noesy Spectra and Three-Dimensional Protein Structure Determination Authors: Mumenthaler, C. / Guntert, P. / Braun, W. / Wuthrich, K. #3:  Journal: Biological Magnetic Resonance / Year: 1999 Journal: Biological Magnetic Resonance / Year: 1999Title: Combined Automated Assignment of NMR Spectra and Calculation of Three- Dimensional Protein Structures Authors: Xu, Y. / Schein, C.H. / Braun, W. #4:  Journal: Biopolymers / Year: 1990 Journal: Biopolymers / Year: 1990Title: A Program, Fantom, for Energy Refinement of Polypeptides and Proteins Using a Newton-Raphson Minimizer in the Torsion Angle Space Authors: Schaumann, T.H. / Braun, W. / Wuthrich, K. #5:  Journal: J.Comput.Chem. / Year: 1998 Journal: J.Comput.Chem. / Year: 1998Title: Exact and Efficient Analytical Calculation of the Accessible Surface Areas and Their Gradients Macromolecules Authors: Fraczkiewicz, R. / Braun, W. | ||||||
| History |
|
- Structure visualization
Structure visualization
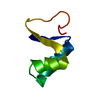
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1cxr.cif.gz 1cxr.cif.gz | 131.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1cxr.ent.gz pdb1cxr.ent.gz | 107.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1cxr.json.gz 1cxr.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/cx/1cxr https://data.pdbj.org/pub/pdb/validation_reports/cx/1cxr ftp://data.pdbj.org/pub/pdb/validation_reports/cx/1cxr ftp://data.pdbj.org/pub/pdb/validation_reports/cx/1cxr | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | Mass: 4728.410 Da / Num. of mol.: 1 / Fragment: ISOFORM OF CRAMBIN (46 RESIDUES) / Mutation: S22P, I25L / Source method: isolated from a natural source Source: (natural)  Crambe hispanica subsp. abyssinica (Abyssinian crambe) Crambe hispanica subsp. abyssinica (Abyssinian crambe)Organ: SEED / Species: Crambe hispanica / Strain: subsp. abyssinica / References: UniProt: P01542 |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||
| NMR details | Text: THIS STRUCTURE WAS DETERMINED USING STANDARD 2D HOMONUCLEAR TECHNIQUES. |
- Sample preparation
Sample preparation
| Details | Contents: 2.5 MM CRAMBIN(SER/ILE) 0.7 ML OF 75% D6-ACETONE, 20%H2O,5%D2O BUFFER |
|---|---|
| Sample conditions | pH: 6.5 / Pressure: 1 atm / Temperature: 298 K |
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer | Type: Varian VXRS / Manufacturer: Varian / Model: VXRS / Field strength: 600 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: SELF-CORRECTING DISTANCE GEOMETRY / Software ordinal: 1 / Details: SEE REFERENCE: J. MAGN. RESON. 136, P76-85(1999) | ||||||||||||||||
| NMR representative | Selection criteria: closest to the average | ||||||||||||||||
| NMR ensemble | Conformer selection criteria: STRUCTURES WITH THE LEAST RESTRAINT VIOLATIONS,STRUCTURES WITH THE LOWEST ENERGY,TARGET FUNCTION Conformers calculated total number: 40 / Conformers submitted total number: 10 |
 Movie
Movie Controller
Controller








 PDBj
PDBj
