[English] 日本語
 Yorodumi
Yorodumi- PDB-1chc: STRUCTURE OF THE C3HC4 DOMAIN BY 1H-NUCLEAR MAGNETIC RESONANCE SP... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1chc | ||||||
|---|---|---|---|---|---|---|---|
| Title | STRUCTURE OF THE C3HC4 DOMAIN BY 1H-NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY; A NEW STRUCTURAL CLASS OF ZINC-FINGER | ||||||
 Components Components | EQUINE HERPES VIRUS-1 RING DOMAIN | ||||||
 Keywords Keywords | VIRAL PROTEIN | ||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated perturbation of host exit from mitosis / symbiont-mediated disruption of host cell PML body / protein monoubiquitination / RING-type E3 ubiquitin transferase / protein polyubiquitination / ubiquitin protein ligase activity / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of IRF3 activity / symbiont-mediated perturbation of host ubiquitin-like protein modification / DNA binding / zinc ion binding Similarity search - Function | ||||||
| Biological species |  Equid herpesvirus 1 (Equine herpesvirus 1) Equid herpesvirus 1 (Equine herpesvirus 1) | ||||||
| Method | SOLUTION NMR | ||||||
 Authors Authors | Barlow, P.N. / Everett, R.D. / Luisi, B. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 1994 Journal: J.Mol.Biol. / Year: 1994Title: Structure of the C3HC4 domain by 1H-nuclear magnetic resonance spectroscopy. A new structural class of zinc-finger. Authors: Barlow, P.N. / Luisi, B. / Milner, A. / Elliott, M. / Everett, R. #1:  Journal: J.Mol.Biol. / Year: 1993 Journal: J.Mol.Biol. / Year: 1993Title: A Novel Arrangement of Zinc-Binding Residues and Secondary Structure in the C3Hc4 Motif of an Alpha Herpes Virus Protein Family Authors: Everett, R.D. / Barlow, P. / Milner, A. / Luisi, B. / Orr, A. / Hope, G. / Lyon, D. #2:  Journal: Cell(Cambridge,Mass.) / Year: 1991 Journal: Cell(Cambridge,Mass.) / Year: 1991Title: A Novel Cysteine-Rich Sequence Motif Authors: Freemont, P.S. / Handon, I.M. / Trowsdale, J. | ||||||
| History |
|
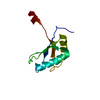
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1chc.cif.gz 1chc.cif.gz | 33.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1chc.ent.gz pdb1chc.ent.gz | 21.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1chc.json.gz 1chc.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ch/1chc https://data.pdbj.org/pub/pdb/validation_reports/ch/1chc ftp://data.pdbj.org/pub/pdb/validation_reports/ch/1chc ftp://data.pdbj.org/pub/pdb/validation_reports/ch/1chc | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| Atom site foot note | 1: CIS PROLINE - PRO 23 | |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 7651.879 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Equid herpesvirus 1 (Equine herpesvirus 1) Equid herpesvirus 1 (Equine herpesvirus 1)Genus: Varicellovirus / References: UniProt: P28990 |
|---|---|
| #2: Chemical |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR |
|---|
- Sample preparation
Sample preparation
| Crystal grow | *PLUS Method: other / Details: NMR |
|---|
- Processing
Processing
| NMR ensemble | Conformers submitted total number: 1 |
|---|
 Movie
Movie Controller
Controller









 PDBj
PDBj



