+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1ccr | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | STRUCTURE OF RICE FERRICYTOCHROME C AT 2.0 ANGSTROMS RESOLUTION | |||||||||
 Components Components | CYTOCHROME C | |||||||||
 Keywords Keywords | ELECTRON TRANSPORT(CYTOCHROME) | |||||||||
| Function / homology |  Function and homology information Function and homology information: / mitochondrial electron transport, cytochrome c to oxygen / mitochondrial electron transport, ubiquinol to cytochrome c / mitochondrial intermembrane space / electron transfer activity / heme binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method |  X-RAY DIFFRACTION / Resolution: 1.5 Å X-RAY DIFFRACTION / Resolution: 1.5 Å | |||||||||
 Authors Authors | Ochi, H. / Hata, Y. / Tanaka, N. / Kakudo, M. / Sakurai, T. / Aihara, S. / Morita, Y. | |||||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 1983 Journal: J.Mol.Biol. / Year: 1983Title: Structure of rice ferricytochrome c at 2.0 A resolution. Authors: Ochi, H. / Hata, Y. / Tanaka, N. / Kakudo, M. / Sakurai, T. / Aihara, S. / Morita, Y. #1:  Journal: J.Biochem.(Tokyo) / Year: 1980 Journal: J.Biochem.(Tokyo) / Year: 1980Title: Amino Acid Sequence of Cytochrome C from Rice Authors: Mori, E. / Morita, Y. #2:  Journal: J.Mol.Biol. / Year: 1972 Journal: J.Mol.Biol. / Year: 1972Title: A Preliminary Crystallographic Investigation of Rice Cytochrome C Authors: Morita, Y. / Ida, S. #3:  Journal: Agric.Biol.Chem. / Year: 1968 Journal: Agric.Biol.Chem. / Year: 1968Title: Studies on Respiratory Enzymes in Rice Kernel. Part I. Isolation and Purification of Cytochrome C and Peroxidase 556 from Rice Embryo Authors: Mori, Y. / Ida, S. | |||||||||
| History |
|
- Structure visualization
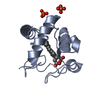
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1ccr.cif.gz 1ccr.cif.gz | 37.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1ccr.ent.gz pdb1ccr.ent.gz | 24.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1ccr.json.gz 1ccr.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1ccr_validation.pdf.gz 1ccr_validation.pdf.gz | 808.6 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1ccr_full_validation.pdf.gz 1ccr_full_validation.pdf.gz | 811.6 KB | Display | |
| Data in XML |  1ccr_validation.xml.gz 1ccr_validation.xml.gz | 8.1 KB | Display | |
| Data in CIF |  1ccr_validation.cif.gz 1ccr_validation.cif.gz | 10 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/cc/1ccr https://data.pdbj.org/pub/pdb/validation_reports/cc/1ccr ftp://data.pdbj.org/pub/pdb/validation_reports/cc/1ccr ftp://data.pdbj.org/pub/pdb/validation_reports/cc/1ccr | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Atom site foot note | 1: RESIDUE LYS 80 AND TML 2 FORM EPSILON-N-TRIMETHYLLYSINE. SEE REMARK 4 ABOVE. 2: RESIDUE LYS 94 AND TML 3 FORM EPSILON-N-TRIMETHYLLYSINE. SEE REMARK 4 ABOVE. |
- Components
Components
| #1: Protein | Mass: 12267.986 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  |
|---|---|
| #2: Chemical | ChemComp-HEC / |
| #3: Water | ChemComp-HOH / |
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.5 Å3/Da / Density % sol: 50.74 % | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | *PLUS pH: 6 / Method: unknown | |||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Radiation | Scattering type: x-ray |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| Reflection | *PLUS Highest resolution: 2 Å / Num. obs: 44000 / % possible obs: 97.5 % / Rmerge(I) obs: 0.022 |
- Processing
Processing
| Refinement | Rfactor Rwork: 0.19 / Highest resolution: 1.5 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement step | Cycle: LAST / Highest resolution: 1.5 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Highest resolution: 2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller












 PDBj
PDBj










