+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: PDB / ID: 1cbh | ||||||
|---|---|---|---|---|---|---|---|
| タイトル | DETERMINATION OF THE THREE-DIMENSIONAL STRUCTURE OF THE C-TERMINAL DOMAIN OF CELLOBIOHYDROLASE I FROM TRICHODERMA REESEI. A STUDY USING NUCLEAR MAGNETIC RESONANCE AND HYBRID DISTANCE GEOMETRY-DYNAMICAL SIMULATED ANNEALING | ||||||
 要素 要素 | C-TERMINAL DOMAIN OF CELLOBIOHYDROLASE I | ||||||
 キーワード キーワード | HYDROLASE (O-GLYCOSYL) | ||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報cellulose 1,4-beta-cellobiosidase (non-reducing end) / cellulose 1,4-beta-cellobiosidase activity / cellulose binding / cellulose catabolic process / extracellular region 類似検索 - 分子機能 | ||||||
| 生物種 |  Hypocrea jecorina (菌類) Hypocrea jecorina (菌類) | ||||||
| 手法 | 溶液NMR | ||||||
 データ登録者 データ登録者 | Clore, G.M. / Gronenborn, A.M. | ||||||
 引用 引用 |  ジャーナル: Biochemistry / 年: 1989 ジャーナル: Biochemistry / 年: 1989タイトル: Determination of the three-dimensional solution structure of the C-terminal domain of cellobiohydrolase I from Trichoderma reesei. A study using nuclear magnetic resonance and hybrid ...タイトル: Determination of the three-dimensional solution structure of the C-terminal domain of cellobiohydrolase I from Trichoderma reesei. A study using nuclear magnetic resonance and hybrid distance geometry-dynamical simulated annealing. 著者: Kraulis, J. / Clore, G.M. / Nilges, M. / Jones, T.A. / Pettersson, G. / Knowles, J. / Gronenborn, A.M. | ||||||
| 履歴 |
|
- 構造の表示
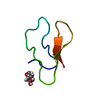
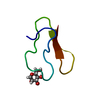
構造の表示
| 構造ビューア | 分子:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- ダウンロードとリンク
ダウンロードとリンク
- ダウンロード
ダウンロード
| PDBx/mmCIF形式 |  1cbh.cif.gz 1cbh.cif.gz | 20.5 KB | 表示 |  PDBx/mmCIF形式 PDBx/mmCIF形式 |
|---|---|---|---|---|
| PDB形式 |  pdb1cbh.ent.gz pdb1cbh.ent.gz | 13 KB | 表示 |  PDB形式 PDB形式 |
| PDBx/mmJSON形式 |  1cbh.json.gz 1cbh.json.gz | ツリー表示 |  PDBx/mmJSON形式 PDBx/mmJSON形式 | |
| その他 |  その他のダウンロード その他のダウンロード |
-検証レポート
| アーカイブディレクトリ |  https://data.pdbj.org/pub/pdb/validation_reports/cb/1cbh https://data.pdbj.org/pub/pdb/validation_reports/cb/1cbh ftp://data.pdbj.org/pub/pdb/validation_reports/cb/1cbh ftp://data.pdbj.org/pub/pdb/validation_reports/cb/1cbh | HTTPS FTP |
|---|
-関連構造データ
- リンク
リンク
- 集合体
集合体
| 登録構造単位 | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR アンサンブル |
|
- 要素
要素
| #1: タンパク質・ペプチド | 分子量: 3746.126 Da / 分子数: 1 / 由来タイプ: 組換発現 / 由来: (組換発現)  Hypocrea jecorina (菌類) Hypocrea jecorina (菌類)参照: UniProt: P62694, cellulose 1,4-beta-cellobiosidase (non-reducing end) |
|---|---|
| Has protein modification | Y |
-実験情報
-実験
| 実験 | 手法: 溶液NMR |
|---|
- 試料調製
試料調製
| 結晶化 | *PLUS 手法: other / 詳細: NMR |
|---|
- 解析
解析
| ソフトウェア |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| NMR software | 名称:  XPLOR / 開発者: BRUNGER / 分類: 精密化 XPLOR / 開発者: BRUNGER / 分類: 精密化 | ||||||||
| 精密化 | ソフトェア番号: 1 詳細: REFINEMENT. THE METHOD USED TO DETERMINE AND REFINE THE STRUCTURE IS THE HYBRID METRIC MATRIX DISTANCE GEOMETRY-DYNAMICAL SIMULATED ANNEALING METHOD (M.NILGES, G.M.CLORE, A.M. GRONENBORN, ...詳細: REFINEMENT. THE METHOD USED TO DETERMINE AND REFINE THE STRUCTURE IS THE HYBRID METRIC MATRIX DISTANCE GEOMETRY-DYNAMICAL SIMULATED ANNEALING METHOD (M.NILGES, G.M.CLORE, A.M. GRONENBORN, FEBS LETT. 229, 317-324 (1988)) USING THE PROGRAM XPLOR (A.T. BRUENGER, YALE UNIVERSITY, CT 06511). STRUCTURAL STATISTICS RMS DEVIATION FROM EXPERIMENTAL RESTRAINTS *(1)* RESTRAINT TYPE NUMBER OF RESTRAINTS RMS (ANGSTROMS) ALL 578 0.024 INTERRESIDUE SHORT RANGE 206 0.030 INTERRESIDUE LONG RANGE 137 0.017 INTRARESIDUE 211 0.021 HBOND *(2)* 24 0.019 POTENTIAL ENERGY TERMS TYPE ENERGY (KCAL/MOL) F(NOE) *(3)* 17 F(TOR) *(4)* 0 F(REPEL) *(5)* 34 LENNARD-JONES VAN DER WAALS ENERGY (E(L-J)) CALCULATED USING THE *CHARMM* EMPIRICAL ENERGY FUNCTION IS -118 KCAL/MOL. DEVIATIONS FROM IDEALIZED GEOMETRY *(6)* TYPE TOTAL NUMBER RMS DEVIATION BONDS 503 0.010 (ANGSTROMS) ANGLES 896 2.170 (DEGREES) IMPROPERS 227 0.911 (DEGREES) NOTES. *(1)* THE RMS DEVIATION FROM THE EXPERIMENTAL RESTRAINTS ARE CALCULATED WITH RESPECT TO THE UPPER AND LOWER LIMITS OF THE DISTANCE RESTRAINTS. NONE OF THE STRUCTURES EXHIBITED VIOLATIONS GREATER THAN 0.5 ANGSTROMS. *(2)* FOR EACH BACKBONE HYDROGEN BOND THERE ARE TWO RESTRAINTS - R(NH-O) .LT. 2.3 ANGSTROMS AND R(N-O) .LT. 3.3 ANGSTROMS. THE LOWER LIMITS ARE GIVEN BY THE SUM OF THE VAN DER WAALS RADII OF THE RELEVANT ATOMS. *(3)* THE VALUES OF THE SQUARE-WELL NOE POTENTIAL F(NOE) ARE CALCULATED WITH A FORCE CONSTANT OF 50 KCAL/MOL/ANGSTROM**2. *(4)* THE VALUES OF F(PHI) ARE CALCULATED WITH A FORCE CONSTANT OF 200 KCAL/MOL/RAD**2. F(PHI) IS A SQUARE-WELL DIHEDRAL POTENTIAL WHICH IS USED TO RESTRICT THE RANGES OF 33 PHI ,24 PSI AND 25 CHI1 TORSION ANGLES. *(5)* THE VALUE OF THE VAN DER WAALS REPULSION TERM F(REPEL) IS CALCULATED WITH A FORCE CONSTANT OF 4 KCAL/MOL/ANGSTROM**4 WITH THE HARD SPHERE VAN DER WAALS RADII SET TO 0.8 TIMES THE STANDARD VALUES USED IN THE *CHARMM* EMPIRICAL ENERGY FUNCTION. *(6)* THE IMPROPER TERMS SERVE TO MAINTAIN PLANARITY AND APPROPRIATE CHIRALITY. THEY ALSO MAINTAIN THE PEPTIDE BONDS OF ALL RESIDUES (WITH THE EXCEPTION OF PROLINES) IN THE TRANS CONFORMATION. IN THE DYNAMICAL SIMULATED ANNEALING CALCULATIONS, THE RESTRAINTS FOR THE DISULFIDE BRIDGES ARE INCLUDED IN THE BOND AND ANGLE TERMS. A TOTAL OF 41 STRUCTURES CONSISTENT WITH THE NMR DATA WERE CALCULATED. THIS ENTRY REPRESENTS THE COORDINATES OBTAINED BY AVERAGING THE COORDINATES OF THE INDIVIDUAL STRUCTURES AND SUBJECTING THE RESULTING COORDINATES TO FURTHER RESTRAINED MINIMIZATION. THE COORDINATES OF THE 41 STRUCTURES ARE GIVEN IN THE PROTEIN DATA BANK ENTRY *2CBH*. THE THERMAL PARAMETERS GIVEN IN THIS ENTRY REPRESENT THE ATOMIC RMS DEVIATION OF THE INDIVIDUAL STRUCTURES ABOUT THE MEAN COORDINATE POSITIONS. ALL THE INTERPROTON DISTANCE AND TORSION ANGLE RESTRAINTS ARE AVAILABLE IN THE RESTRAINT FILE. | ||||||||
| NMRアンサンブル | 登録したコンフォーマーの数: 1 |
 ムービー
ムービー コントローラー
コントローラー












 PDBj
PDBj