[English] 日本語
 Yorodumi
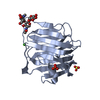
Yorodumi- PDB-1c4r: THE STRUCTURE OF THE LIGAND-BINDING DOMAIN OF NEUREXIN 1BETA: REG... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1c4r | ||||||
|---|---|---|---|---|---|---|---|
| Title | THE STRUCTURE OF THE LIGAND-BINDING DOMAIN OF NEUREXIN 1BETA: REGULATION OF LNS DOMAIN FUNCTION BY ALTERNATIVE SPLICING | ||||||
 Components Components | NEUREXIN-I BETA | ||||||
 Keywords Keywords | MEMBRANE PROTEIN / LECTIN-LIKE / NEUROBIOLOGY / CELL-CELL ADHESION / CELL-CELL RECOGNITION / ALTERNATIVE SPLICING | ||||||
| Function / homology |  Function and homology information Function and homology informationprotein-containing complex assembly involved in synapse maturation / positive regulation of presynaptic active zone assembly / cell-cell adhesion involved in synapse maturation / : / protein complex involved in cell-cell adhesion / guanylate kinase-associated protein clustering / positive regulation of neuromuscular synaptic transmission / neuron to neuron synapse / neuroligin clustering involved in postsynaptic membrane assembly / regulation of trans-synaptic signaling by endocannabinoid, modulating synaptic transmission ...protein-containing complex assembly involved in synapse maturation / positive regulation of presynaptic active zone assembly / cell-cell adhesion involved in synapse maturation / : / protein complex involved in cell-cell adhesion / guanylate kinase-associated protein clustering / positive regulation of neuromuscular synaptic transmission / neuron to neuron synapse / neuroligin clustering involved in postsynaptic membrane assembly / regulation of trans-synaptic signaling by endocannabinoid, modulating synaptic transmission / trans-synaptic signaling, modulating synaptic transmission / trans-synaptic protein complex / type 1 fibroblast growth factor receptor binding / negative regulation of filopodium assembly / gephyrin clustering involved in postsynaptic density assembly / cerebellar granule cell differentiation / slit diaphragm / postsynaptic density protein 95 clustering / postsynaptic membrane assembly / synapse maturation / gamma-aminobutyric acid receptor clustering / presynaptic membrane assembly / neuroligin family protein binding / vocal learning / positive regulation of synapse maturation / regulation of grooming behavior / presynapse assembly / maintenance of synapse structure / synaptic vesicle clustering / synaptic membrane adhesion / regulation of postsynaptic specialization assembly / positive regulation of fibroblast growth factor receptor signaling pathway / receptor localization to synapse / neuron cell-cell adhesion / NMDA glutamate receptor clustering / inhibitory synapse / calcium-dependent cell-cell adhesion / vocalization behavior / regulation of postsynaptic density assembly / protein localization to synapse / acetylcholine receptor binding / regulation of synaptic vesicle cycle / AMPA selective glutamate receptor signaling pathway / neurotransmitter secretion / positive regulation of synapse assembly / NMDA selective glutamate receptor signaling pathway / heterophilic cell-cell adhesion / neuromuscular process controlling balance / positive regulation of phospholipase C-activating G protein-coupled receptor signaling pathway / adult behavior / social behavior / endocytic vesicle / regulation of presynapse assembly / prepulse inhibition / positive regulation of synaptic transmission, glutamatergic / axonal growth cone / synapse assembly / neuron projection morphogenesis / cell adhesion molecule binding / presynaptic active zone membrane / excitatory synapse / positive regulation of excitatory postsynaptic potential / cellular response to calcium ion / positive regulation of synaptic transmission, GABAergic / learning / positive regulation of protein localization to plasma membrane / calcium channel regulator activity / neuromuscular junction / establishment of protein localization / positive regulation of neuron projection development / circadian rhythm / GABA-ergic synapse / Schaffer collateral - CA1 synapse / neuron projection development / calcium-dependent protein binding / transmembrane signaling receptor activity / presynaptic membrane / angiogenesis / nuclear membrane / vesicle / chemical synaptic transmission / positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / positive regulation of ERK1 and ERK2 cascade / signaling receptor binding / negative regulation of gene expression / neuronal cell body / calcium ion binding / positive regulation of gene expression / protein-containing complex binding / glutamatergic synapse / cell surface / endoplasmic reticulum / signal transduction / protein-containing complex / plasma membrane Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / Resolution: 2.6 Å SYNCHROTRON / Resolution: 2.6 Å | ||||||
 Authors Authors | Rudenko, G. / Nguyen, T. / Chelliah, Y. / Sudhof, T.C. / Deisenhofer, J. | ||||||
 Citation Citation |  Journal: Cell(Cambridge,Mass.) / Year: 1999 Journal: Cell(Cambridge,Mass.) / Year: 1999Title: The structure of the ligand-binding domain of neurexin Ibeta: regulation of LNS domain function by alternative splicing. Authors: Rudenko, G. / Nguyen, T. / Chelliah, Y. / Sudhof, T.C. / Deisenhofer, J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1c4r.cif.gz 1c4r.cif.gz | 275.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1c4r.ent.gz pdb1c4r.ent.gz | 226.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1c4r.json.gz 1c4r.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/c4/1c4r https://data.pdbj.org/pub/pdb/validation_reports/c4/1c4r ftp://data.pdbj.org/pub/pdb/validation_reports/c4/1c4r ftp://data.pdbj.org/pub/pdb/validation_reports/c4/1c4r | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||||||||||||||||||||||
| 2 | 
| ||||||||||||||||||||||||||||||||
| 3 | 
| ||||||||||||||||||||||||||||||||
| 4 | 
| ||||||||||||||||||||||||||||||||
| 5 | 
| ||||||||||||||||||||||||||||||||
| 6 | 
| ||||||||||||||||||||||||||||||||
| 7 | 
| ||||||||||||||||||||||||||||||||
| 8 | 
| ||||||||||||||||||||||||||||||||
| Unit cell |
| ||||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS oper:
|
- Components
Components
| #1: Protein | Mass: 19693.072 Da / Num. of mol.: 8 / Fragment: EXTRACELLULAR DOMAIN Source method: isolated from a genetically manipulated source Source: (gene. exp.)   #2: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 2 X-RAY DIFFRACTION / Number of used crystals: 2 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.75 Å3/Da / Density % sol: 67.24 % | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | pH: 6.5 / Details: pH 6.50 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 21 ℃ / pH: 7.5 / Method: vapor diffusion | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 110 K | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SSRL SSRL  / Beamline: BL1-5 / Wavelength: 1.0712, 0.9791, 0.9793, 0.9221 / Beamline: BL1-5 / Wavelength: 1.0712, 0.9791, 0.9793, 0.9221 | |||||||||||||||
| Radiation | Protocol: MAD / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||||||||
| Radiation wavelength |
| |||||||||||||||
| Reflection | Biso Wilson estimate: 29.3 Å2 | |||||||||||||||
| Reflection | *PLUS Highest resolution: 2.6 Å / Lowest resolution: 20 Å / Num. obs: 73128 / % possible obs: 98.1 % / Num. measured all: 351083 / Rmerge(I) obs: 0.095 | |||||||||||||||
| Reflection shell | *PLUS Highest resolution: 2.6 Å / Lowest resolution: 2.64 Å / % possible obs: 82.7 % / Rmerge(I) obs: 0.495 / Mean I/σ(I) obs: 3.1 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 2.6→20 Å / Rfactor Rfree error: 0.005 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: FLAT MODEL / Bsol: 38.87 Å2 / ksol: 0.37 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 38.2 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.6→20 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints NCS | NCS model details: CONSTR | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.6→2.76 Å / Rfactor Rfree error: 0.018 / Total num. of bins used: 6
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name: CNS / Version: 0.5 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller










 PDBj
PDBj

