[English] 日本語
 Yorodumi
Yorodumi- PDB-1c34: SOLUTION STRUCTURE OF A QUADRUPLEX FORMING DNA AND ITS INTERMIDIATE -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1c34 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | SOLUTION STRUCTURE OF A QUADRUPLEX FORMING DNA AND ITS INTERMIDIATE | ||||||||||||||||||
 Components Components | DNA (5'-D(* Keywords KeywordsDNA / APTAMER / THROMBIN BINDING / POTASSIUM / METAL BINDING SITES / QUADRUPLEX / INTERMEDIATE | Function / homology | DNA / DNA (> 10) |  Function and homology information Function and homology informationMethod | SOLUTION NMR / STRUCTURE REFINEMENT OF THE DNA WAS PREFORMED USING COMPLETE RELAXATION MATRIX, RESTRAINED MOLECULAR DYNAMICS. |  Authors AuthorsBolton, P.H. / Marathias, V.M. / Wang, K. |  Citation Citation Journal: Nucleic Acids Res. / Year: 2000 Journal: Nucleic Acids Res. / Year: 2000Title: Structures of the potassium-saturated, 2:1, and intermediate, 1:1, forms of a quadruplex DNA. Authors: Marathias, V.M. / Bolton, P.H. #1:  Journal: J.Mol.Biol. / Year: 1996 Journal: J.Mol.Biol. / Year: 1996Title: Determination of the Number and Location of the Manganese Binding Sites of DNA Quadruplexes in Solution by EPR and NMR in the Presence and Absence of Thrombin Authors: Bolton, P.H. / Marathias, V.M. / Wang, K. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1c34.cif.gz 1c34.cif.gz | 121.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1c34.ent.gz pdb1c34.ent.gz | 95.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1c34.json.gz 1c34.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/c3/1c34 https://data.pdbj.org/pub/pdb/validation_reports/c3/1c34 ftp://data.pdbj.org/pub/pdb/validation_reports/c3/1c34 ftp://data.pdbj.org/pub/pdb/validation_reports/c3/1c34 | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
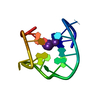
| Deposited unit | 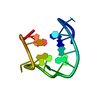
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: DNA chain | Mass: 4743.051 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: THROMBIN BINDING APTAMER INTERMIDIATE |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||||||
| NMR details | Text: THIS STRUCTURE IS THE INTERMEDIATE FORM OF THE 15MER QUADRUPLEX WITH ONLY 1 POTASSSIUM PER DNA MOLECULE. |
- Sample preparation
Sample preparation
| Details |
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample conditions |
| |||||||||||||||
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: STRUCTURE REFINEMENT OF THE DNA WAS PREFORMED USING COMPLETE RELAXATION MATRIX, RESTRAINED MOLECULAR DYNAMICS. Software ordinal: 1 | ||||||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: 80 TO 100PS STRUCTURES FROM DYNAMICS Conformers calculated total number: 11 / Conformers submitted total number: 11 |
 Movie
Movie Controller
Controller










 PDBj
PDBj






































 X-PLOR
X-PLOR