+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1b3p | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | 5'-D(*GP*GP*AP*GP*GP*AP*T)-3' | ||||||||||||||||||
 Components Components | DNA (5'-D(* Keywords KeywordsDNA / (G-G-A) TRIPLET REPEAT / V-SHAPED BACKBONE / PARALLEL-STRANDED SEGMENTS / MISMATCH ALIGNMENTS / UNIFORM 13C / 15N-LABELED DNA | Function / homology | DNA |  Function and homology information Function and homology informationMethod | SOLUTION NMR / distance geometry |  Authors AuthorsKettani, A. / Bouaziz, S. / Skripkin, E. / Majumdar, A. / Wang, W. / Jones, R.A. / Patel, D.J. |  Citation Citation Journal: Structure Fold.Des. / Year: 1999 Journal: Structure Fold.Des. / Year: 1999Title: Interlocked mismatch-aligned arrowhead DNA motifs. Authors: Kettani, A. / Bouaziz, S. / Skripkin, E. / Majumdar, A. / Wang, W. / Jones, R.A. / Patel, D.J. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1b3p.cif.gz 1b3p.cif.gz | 151.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1b3p.ent.gz pdb1b3p.ent.gz | 122.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1b3p.json.gz 1b3p.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/b3/1b3p https://data.pdbj.org/pub/pdb/validation_reports/b3/1b3p ftp://data.pdbj.org/pub/pdb/validation_reports/b3/1b3p ftp://data.pdbj.org/pub/pdb/validation_reports/b3/1b3p | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 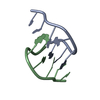
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: DNA chain | Mass: 2202.472 Da / Num. of mol.: 2 / Source method: obtained synthetically |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||||||||||
| NMR details | Text: THE STRUCTURE WAS DETERMINED USING 2D HOMONUCLEAR AND HETERONUCLEAR EXPERIMENTS ON 13C AND 15N LABELED SAMPLE. THE DISTANCE RESTRAINTS WERE DEDUCED FROM 5 NOESY MIXING TIMES (50, 100, 150, 200 ...Text: THE STRUCTURE WAS DETERMINED USING 2D HOMONUCLEAR AND HETERONUCLEAR EXPERIMENTS ON 13C AND 15N LABELED SAMPLE. THE DISTANCE RESTRAINTS WERE DEDUCED FROM 5 NOESY MIXING TIMES (50, 100, 150, 200 AND 250 MS) IN D2O AND 2 NOESY MIXING TIMES IN H2O (60 AND 200 MS) MS |
- Sample preparation
Sample preparation
| Sample conditions | Ionic strength: 10 mM NACL, 2 mM PHOSPHATE / pH: 6.6 / Pressure: ambient / Temperature: 273 K |
|---|---|
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer | Type: Varian UNITYPLUS / Manufacturer: Varian / Model: UNITYPLUS / Field strength: 600 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: distance geometry / Software ordinal: 1 Details: REFINEMENT DETAILS CAN BE FOUND IN THE JNRL CITATION ABOVE | ||||||||||||
| NMR ensemble | Conformer selection criteria: ACCEPTABLE COVALENT GEOMETRY, LOW DISTANCE RESTRAINTS VIIOLATIONS AND FAVORABLE NON-BONDED ENERGY VALUES Conformers calculated total number: 100 / Conformers submitted total number: 15 |
 Movie
Movie Controller
Controller








 PDBj
PDBj

 X-PLOR
X-PLOR