+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1atg | ||||||
|---|---|---|---|---|---|---|---|
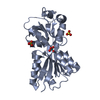
| Title | AZOTOBACTER VINELANDII PERIPLASMIC MOLYBDATE-BINDING PROTEIN | ||||||
 Components Components | PERIPLASMIC MOLYBDATE-BINDING PROTEIN | ||||||
 Keywords Keywords | BINDING PROTEIN / MOLYBDATE / TUNGSTATE / PERIPLASM / ABC TRANSPORTER | ||||||
| Function / homology |  Function and homology information Function and homology information | ||||||
| Biological species |  Azotobacter vinelandii (bacteria) Azotobacter vinelandii (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SINGLE ISOMORPHOUS REPLACEMENT WITH ANOMALOUS SCATTERING / Resolution: 1.2 Å SYNCHROTRON / SINGLE ISOMORPHOUS REPLACEMENT WITH ANOMALOUS SCATTERING / Resolution: 1.2 Å | ||||||
 Authors Authors | Lawson, D.M. / Pau, R.N. / Williams, C.E.M. / Mitchenall, L.A. | ||||||
 Citation Citation |  Journal: Structure / Year: 1998 Journal: Structure / Year: 1998Title: Ligand size is a major determinant of specificity in periplasmic oxyanion-binding proteins: the 1.2 A resolution crystal structure of Azotobacter vinelandii ModA. Authors: Lawson, D.M. / Williams, C.E. / Mitchenall, L.A. / Pau, R.N. #1:  Journal: J.Chem.Soc.,Dalton Trans. / Year: 1997 Journal: J.Chem.Soc.,Dalton Trans. / Year: 1997Title: Protein Ligands for Molybdate. Specificity and Charge Stabilisation at the Anion-Binding Sites of Periplasmic and Intracellular Molybdate-Binding Proteins of Azotobacter Vinelandii Authors: Lawson, D.M. / Williams, C.E.M. / White, D.J. / Choay, A.P. / Mitchenall, L.A. / Pau, R.N. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1atg.cif.gz 1atg.cif.gz | 65.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1atg.ent.gz pdb1atg.ent.gz | 47 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1atg.json.gz 1atg.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/at/1atg https://data.pdbj.org/pub/pdb/validation_reports/at/1atg ftp://data.pdbj.org/pub/pdb/validation_reports/at/1atg ftp://data.pdbj.org/pub/pdb/validation_reports/at/1atg | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 24390.729 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Azotobacter vinelandii (bacteria) / Cellular location: PERIPLASM / Strain: RP2 / References: UniProt: Q7SIH2 Azotobacter vinelandii (bacteria) / Cellular location: PERIPLASM / Strain: RP2 / References: UniProt: Q7SIH2 |
|---|
-Non-polymers , 5 types, 360 molecules 








| #2: Chemical | ChemComp-WO4 / | ||
|---|---|---|---|
| #3: Chemical | ChemComp-ACT / | ||
| #4: Chemical | ChemComp-SO4 / | ||
| #5: Chemical | | #6: Water | ChemComp-HOH / | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.54 Å3/Da / Density % sol: 51 % | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 291 K / Method: vapor diffusion, hanging drop / pH: 4 Details: HANGING DROP VAPOUR DIFFUSION WITH PROTEIN CONCENTRATION IN THE RANGE 10-15 MG/ML IN 10MM TRIS-HCL PH 7.0. 1 OR 2 MICROLITER DROPS OF THE LATTER WERE MIXED WITH AN EQUAL VOLUME OF 11-14% ...Details: HANGING DROP VAPOUR DIFFUSION WITH PROTEIN CONCENTRATION IN THE RANGE 10-15 MG/ML IN 10MM TRIS-HCL PH 7.0. 1 OR 2 MICROLITER DROPS OF THE LATTER WERE MIXED WITH AN EQUAL VOLUME OF 11-14% (W/V) PEG 4000 AND 0.4M AMMONIUM SULFATE IN 0.1M ACETATE BUFFER AT PH 4.0, AND THEN EQUILIBRATED AGAINST THIS SOLUTION AT 18 DEG C. CRYOPROTECTED USING THE SAME SOLUTION CONTAINING 25% (V/V) ETHYLENE GLYCOL., vapor diffusion - hanging drop, temperature 291K PH range: 4.0-7.0 | ||||||||||||||||||||||||||||||||||||||||||
| Crystal | *PLUS | ||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 18 ℃ / pH: 7 / Method: vapor diffusion, hanging drop | ||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  EMBL/DESY, HAMBURG EMBL/DESY, HAMBURG  / Beamline: X11 / Wavelength: 0.9116 / Beamline: X11 / Wavelength: 0.9116 |
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE / Date: Sep 1, 1996 / Details: MIRRORS |
| Radiation | Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9116 Å / Relative weight: 1 |
| Reflection | Resolution: 1.2→100 Å / Num. obs: 74061 / % possible obs: 99.4 % / Observed criterion σ(I): 0 / Redundancy: 3.7 % / Rmerge(I) obs: 0.094 / Net I/σ(I): 12.7 |
| Reflection shell | Resolution: 1.2→1.22 Å / Redundancy: 3.2 % / Rmerge(I) obs: 0.161 / Mean I/σ(I) obs: 6.9 / % possible all: 99.1 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure: SINGLE ISOMORPHOUS REPLACEMENT WITH ANOMALOUS SCATTERING Resolution: 1.2→40 Å / Cross valid method: THROUGHOUT / σ(F): 0 Details: LUZZATI ESD BASED ON TEST REFLECTIONS ONLY. PLANE RESTRAINT RMS BASED ON NON-AROMATIC PLANAR GROUPS ONLY.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.13 Å / Luzzati d res low obs: 5 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.2→40 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name: REFMAC / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Rfactor obs: 0.164 / Rfactor Rfree: 0.18354 / Rfactor Rwork: 0.1644 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller












 PDBj
PDBj



