[English] 日本語
 Yorodumi
Yorodumi- PDB-1arc: THE PRIMARY STRUCTURE AND STRUCTURAL CHARACTERISTICS OF ACHROMOBA... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1arc | ||||||
|---|---|---|---|---|---|---|---|
| Title | THE PRIMARY STRUCTURE AND STRUCTURAL CHARACTERISTICS OF ACHROMOBACTER LYTICUS PROTEASE I, A LYSINE-SPECIFIC SERINE PROTEASE | ||||||
 Components Components | ACHROMOBACTER PROTEASE I | ||||||
 Keywords Keywords | HYDROLASE/HYDROLASE INHIBITOR / SERINE PROTEASE / HYDROLASE-HYDROLASE INHIBITOR complex | ||||||
| Function / homology |  Function and homology information Function and homology informationlysyl endopeptidase / serine-type endopeptidase activity / proteolysis / extracellular region Similarity search - Function | ||||||
| Biological species |  Achromobacter lyticus (bacteria) Achromobacter lyticus (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / Resolution: 2 Å X-RAY DIFFRACTION / Resolution: 2 Å | ||||||
 Authors Authors | Kitagawa, Y. / Katsube, Y. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 1989 Journal: J.Biol.Chem. / Year: 1989Title: The primary structure and structural characteristics of Achromobacter lyticus protease I, a lysine-specific serine protease. Authors: Tsunasawa, S. / Masaki, T. / Hirose, M. / Soejima, M. / Sakiyama, F. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1arc.cif.gz 1arc.cif.gz | 62 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1arc.ent.gz pdb1arc.ent.gz | 44.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1arc.json.gz 1arc.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1arc_validation.pdf.gz 1arc_validation.pdf.gz | 436.4 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1arc_full_validation.pdf.gz 1arc_full_validation.pdf.gz | 439.9 KB | Display | |
| Data in XML |  1arc_validation.xml.gz 1arc_validation.xml.gz | 7.2 KB | Display | |
| Data in CIF |  1arc_validation.cif.gz 1arc_validation.cif.gz | 10.7 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ar/1arc https://data.pdbj.org/pub/pdb/validation_reports/ar/1arc ftp://data.pdbj.org/pub/pdb/validation_reports/ar/1arc ftp://data.pdbj.org/pub/pdb/validation_reports/ar/1arc | HTTPS FTP |
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 27759.227 Da / Num. of mol.: 1 / Fragment: residues 206-473 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Achromobacter lyticus (bacteria) / References: UniProt: P15636, lysyl endopeptidase Achromobacter lyticus (bacteria) / References: UniProt: P15636, lysyl endopeptidase |
|---|---|
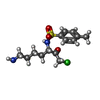
| #2: Chemical | ChemComp-TCK / |
| #3: Water | ChemComp-HOH / |
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.15 Å3/Da / Density % sol: 42.75 % |
|---|---|
| Crystal grow | *PLUS Method: other |
-Data collection
| Radiation | Scattering type: x-ray |
|---|---|
| Radiation wavelength | Relative weight: 1 |
- Processing
Processing
| Software | Name: PROLSQ / Classification: refinement | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Rfactor obs: 0.152 / Highest resolution: 2 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Highest resolution: 2 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Highest resolution: 2 Å / Rfactor obs: 0.152 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller











 PDBj
PDBj