[English] 日本語
 Yorodumi
Yorodumi- PDB-124d: STRUCTURE OF A DNA:RNA HYBRID DUPLEX: WHY RNASE H DOES NOT CLEAVE... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 124d | ||||||
|---|---|---|---|---|---|---|---|
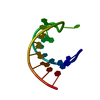
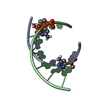
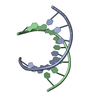
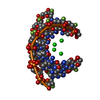
| Title | STRUCTURE OF A DNA:RNA HYBRID DUPLEX: WHY RNASE H DOES NOT CLEAVE PURE RNA | ||||||
 Components Components |
| ||||||
 Keywords Keywords | DNA-RNA HYBRID / DNA-RNA COMPLEX / DOUBLE HELIX | ||||||
| Function / homology | DNA / RNA Function and homology information Function and homology information | ||||||
| Method | SOLUTION NMR / RESTRAINED MOLECULAR DYNAMICS, NOESY SIMULATION REFINEMENT | ||||||
 Authors Authors | Fedoroff, O.Y. / Salazar, M. / Reid, B.R. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 1993 Journal: J.Mol.Biol. / Year: 1993Title: Structure of a DNA:RNA Hybrid Duplex. Why Rnase H Does not Cleave Pure RNA Authors: Fedoroff, O.Y. / Salazar, M. / Reid, B.R. #1:  Journal: Biochemistry / Year: 1993 Journal: Biochemistry / Year: 1993Title: The DNA Strand in DNA-RNA Hybrid Duplexes is Neither B-Form Nor A-Form in Solution Authors: Salazar, M. / Fedoroff, O.Y. / Miller, J.M. / Ribeiro, N.S. / Reid, B.R. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  124d.cif.gz 124d.cif.gz | 19.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb124d.ent.gz pdb124d.ent.gz | 11 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  124d.json.gz 124d.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  124d_validation.pdf.gz 124d_validation.pdf.gz | 293.7 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  124d_full_validation.pdf.gz 124d_full_validation.pdf.gz | 293.5 KB | Display | |
| Data in XML |  124d_validation.xml.gz 124d_validation.xml.gz | 1.6 KB | Display | |
| Data in CIF |  124d_validation.cif.gz 124d_validation.cif.gz | 1.8 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/24/124d https://data.pdbj.org/pub/pdb/validation_reports/24/124d ftp://data.pdbj.org/pub/pdb/validation_reports/24/124d ftp://data.pdbj.org/pub/pdb/validation_reports/24/124d | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: DNA chain | Mass: 2426.617 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: CHEMICALLY SYNTHESIZED |
|---|---|
| #2: RNA chain | Mass: 2526.561 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: CHEMICALLY SYHTHESIZED |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR |
|---|
- Sample preparation
Sample preparation
| Crystal grow | *PLUS Method: other / Details: NMR |
|---|
- Processing
Processing
| Refinement | Method: RESTRAINED MOLECULAR DYNAMICS, NOESY SIMULATION REFINEMENT Software ordinal: 1 Details: 132 NOE RESTRAINTS AND 20 HYDROGEN BONDS WERE USED. |
|---|---|
| NMR ensemble | Conformers calculated total number: 1 / Conformers submitted total number: 1 |
 Movie
Movie Controller
Controller






 PDBj
PDBj