[English] 日本語
 Yorodumi
Yorodumi- EMDB-9844: cryo-EM structure of DOT1L bound to H2B ubiquitinated nucleosome -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9844 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryo-EM structure of DOT1L bound to H2B ubiquitinated nucleosome | ||||||||||||
 Map data Map data | catalytic domain of DOT1L bound to H2B ubiquitinated nucleosome. | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | histone / nucleosome / methylation / GENE REGULATION | ||||||||||||
| Function / homology |  Function and homology information Function and homology information[histone H3]-lysine79 N-trimethyltransferase / histone H3K79 methyltransferase activity / histone H3K79 trimethyltransferase activity / regulation of transcription regulatory region DNA binding / hypothalamus gonadotrophin-releasing hormone neuron development / female meiosis I / positive regulation of protein monoubiquitination / regulation of receptor signaling pathway via JAK-STAT / fat pad development / mitochondrion transport along microtubule ...[histone H3]-lysine79 N-trimethyltransferase / histone H3K79 methyltransferase activity / histone H3K79 trimethyltransferase activity / regulation of transcription regulatory region DNA binding / hypothalamus gonadotrophin-releasing hormone neuron development / female meiosis I / positive regulation of protein monoubiquitination / regulation of receptor signaling pathway via JAK-STAT / fat pad development / mitochondrion transport along microtubule / histone H3 methyltransferase activity / histone methyltransferase activity / female gonad development / seminiferous tubule development / male meiosis I / positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator / subtelomeric heterochromatin formation / energy homeostasis / neuron projection morphogenesis / regulation of proteasomal protein catabolic process / Maturation of protein E / Maturation of protein E / ER Quality Control Compartment (ERQC) / Myoclonic epilepsy of Lafora / FLT3 signaling by CBL mutants / Constitutive Signaling by NOTCH1 HD Domain Mutants / IRAK2 mediated activation of TAK1 complex / Prevention of phagosomal-lysosomal fusion / Alpha-protein kinase 1 signaling pathway / Glycogen synthesis / IRAK1 recruits IKK complex / IRAK1 recruits IKK complex upon TLR7/8 or 9 stimulation / Endosomal Sorting Complex Required For Transport (ESCRT) / Membrane binding and targetting of GAG proteins / Negative regulation of FLT3 / Regulation of TBK1, IKKε (IKBKE)-mediated activation of IRF3, IRF7 / PTK6 Regulates RTKs and Their Effectors AKT1 and DOK1 / Regulation of TBK1, IKKε-mediated activation of IRF3, IRF7 upon TLR3 ligation / IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation / NOTCH2 Activation and Transmission of Signal to the Nucleus / TICAM1,TRAF6-dependent induction of TAK1 complex / TICAM1-dependent activation of IRF3/IRF7 / APC/C:Cdc20 mediated degradation of Cyclin B / telomere organization / Regulation of FZD by ubiquitination / Downregulation of ERBB4 signaling / APC-Cdc20 mediated degradation of Nek2A / p75NTR recruits signalling complexes / InlA-mediated entry of Listeria monocytogenes into host cells / TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling / Regulation of pyruvate metabolism / NF-kB is activated and signals survival / TRAF6-mediated induction of TAK1 complex within TLR4 complex / regulation of neuron apoptotic process / Downregulation of ERBB2:ERBB3 signaling / Pexophagy / Regulation of innate immune responses to cytosolic DNA / NRIF signals cell death from the nucleus / Regulation of PTEN localization / Activated NOTCH1 Transmits Signal to the Nucleus / DNA damage checkpoint signaling / VLDLR internalisation and degradation / positive regulation of protein ubiquitination / Synthesis of active ubiquitin: roles of E1 and E2 enzymes / TICAM1, RIP1-mediated IKK complex recruitment / Regulation of BACH1 activity / Translesion synthesis by REV1 / MAP3K8 (TPL2)-dependent MAPK1/3 activation / Degradation of CDH1 / Translesion synthesis by POLK / InlB-mediated entry of Listeria monocytogenes into host cell / JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 / Activation of IRF3, IRF7 mediated by TBK1, IKKε (IKBKE) / Josephin domain DUBs / Downregulation of TGF-beta receptor signaling / Translesion synthesis by POLI / Gap-filling DNA repair synthesis and ligation in GG-NER / IKK complex recruitment mediated by RIP1 / Degradation of CRY and PER proteins / Regulation of activated PAK-2p34 by proteasome mediated degradation / PINK1-PRKN Mediated Mitophagy / regulation of mitochondrial membrane potential / TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) / TNFR1-induced NF-kappa-B signaling pathway / Autodegradation of Cdh1 by Cdh1:APC/C / TCF dependent signaling in response to WNT / Regulation of NF-kappa B signaling / APC/C:Cdc20 mediated degradation of Securin / N-glycan trimming in the ER and Calnexin/Calreticulin cycle / activated TAK1 mediates p38 MAPK activation / Asymmetric localization of PCP proteins / Ubiquitin-dependent degradation of Cyclin D / SCF-beta-TrCP mediated degradation of Emi1 / NIK-->noncanonical NF-kB signaling / Regulation of signaling by CBL / TNFR2 non-canonical NF-kB pathway / AUF1 (hnRNP D0) binds and destabilizes mRNA / Negative regulators of DDX58/IFIH1 signaling / NOTCH3 Activation and Transmission of Signal to the Nucleus / Assembly of the pre-replicative complex Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
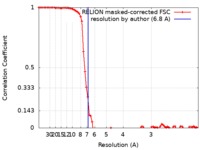
| Method | single particle reconstruction / cryo EM / Resolution: 6.8 Å | ||||||||||||
 Authors Authors | Jang S / Song JJ | ||||||||||||
| Funding support |  Korea, Republic Of, 3 items Korea, Republic Of, 3 items
| ||||||||||||
 Citation Citation |  Journal: Genes Dev / Year: 2019 Journal: Genes Dev / Year: 2019Title: Structural basis of recognition and destabilization of the histone H2B ubiquitinated nucleosome by the DOT1L histone H3 Lys79 methyltransferase. Authors: Seongmin Jang / Chanshin Kang / Han-Sol Yang / Taeyang Jung / Hans Hebert / Ka Young Chung / Seung Joong Kim / Sungchul Hohng / Ji-Joon Song /   Abstract: DOT1L is a histone H3 Lys79 methyltransferase whose activity is stimulated by histone H2B Lys120 ubiquitination, suggesting cross-talk between histone H3 methylation and H2B ubiquitination. Here, we ...DOT1L is a histone H3 Lys79 methyltransferase whose activity is stimulated by histone H2B Lys120 ubiquitination, suggesting cross-talk between histone H3 methylation and H2B ubiquitination. Here, we present cryo-EM structures of DOT1L complexes with unmodified or H2B ubiquitinated nucleosomes, showing that DOT1L recognizes H2B ubiquitin and the H2A/H2B acidic patch through a C-terminal hydrophobic helix and an arginine anchor in DOT1L, respectively. Furthermore, the structures combined with single-molecule FRET experiments show that H2B ubiquitination enhances a noncatalytic function of the DOT1L-destabilizing nucleosome. These results establish the molecular basis of the cross-talk between H2B ubiquitination and H3 Lys79 methylation as well as nucleosome destabilization by DOT1L. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9844.map.gz emd_9844.map.gz | 37.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9844-v30.xml emd-9844-v30.xml emd-9844.xml emd-9844.xml | 17.6 KB 17.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9844_fsc.xml emd_9844_fsc.xml | 8 KB | Display |  FSC data file FSC data file |
| Images |  emd_9844.png emd_9844.png | 112.4 KB | ||
| Filedesc metadata |  emd-9844.cif.gz emd-9844.cif.gz | 6.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9844 http://ftp.pdbj.org/pub/emdb/structures/EMD-9844 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9844 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9844 | HTTPS FTP |
-Related structure data
| Related structure data |  6jmaMC  9843C  6jm9C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9844.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9844.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | catalytic domain of DOT1L bound to H2B ubiquitinated nucleosome. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : DOT1L bound to H2B ubiquitinated nucleosome
| Entire | Name: DOT1L bound to H2B ubiquitinated nucleosome |
|---|---|
| Components |
|
-Supramolecule #1: DOT1L bound to H2B ubiquitinated nucleosome
| Supramolecule | Name: DOT1L bound to H2B ubiquitinated nucleosome / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#7 |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 290 KDa |
-Macromolecule #1: DNA I&J
| Macromolecule | Name: DNA I&J / type: dna / ID: 1 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 35.168547 KDa |
| Sequence | String: (DA)(DG)(DA)(DT)(DT)(DC)(DT)(DA)(DC)(DC) (DA)(DA)(DA)(DA)(DG)(DT)(DG)(DT)(DA)(DT) (DT)(DT)(DG)(DG)(DA)(DA)(DA)(DC)(DT) (DG)(DC)(DT)(DC)(DC)(DA)(DT)(DC)(DA)(DA) (DA) (DA)(DG)(DG)(DC)(DA)(DT) ...String: (DA)(DG)(DA)(DT)(DT)(DC)(DT)(DA)(DC)(DC) (DA)(DA)(DA)(DA)(DG)(DT)(DG)(DT)(DA)(DT) (DT)(DT)(DG)(DG)(DA)(DA)(DA)(DC)(DT) (DG)(DC)(DT)(DC)(DC)(DA)(DT)(DC)(DA)(DA) (DA) (DA)(DG)(DG)(DC)(DA)(DT)(DG)(DT) (DT)(DC)(DA)(DG)(DC)(DT)(DG)(DA)(DA)(DT) (DT)(DC) (DA)(DG)(DC)(DT)(DG)(DA)(DA) (DC)(DA)(DT)(DG)(DC)(DC)(DT)(DT)(DT)(DT) (DG)(DA)(DT) (DG)(DG)(DA)(DG)(DC)(DA) (DG)(DT)(DT)(DT)(DC)(DC)(DA)(DA)(DA)(DT) (DA)(DC)(DA)(DC) (DT)(DT)(DT)(DT)(DG) (DG)(DT)(DA)(DG)(DA)(DA)(DT)(DC)(DT) |
-Macromolecule #2: Histone H3.2
| Macromolecule | Name: Histone H3.2 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 11.48841 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: PHRYRPGTVA LREIRRYQKS TELLIRKLPF QRLVREIAQD FKTDLRFQSS AVMALQEASE AYLVGLFEDT NLCAIHAKRV TIMPKDIQL ARRIRGERA UniProtKB: Histone H3.2 |
-Macromolecule #3: Histone H4
| Macromolecule | Name: Histone H4 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 9.99077 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: KRHRKVLRDN IQGITKPAIR RLARRGGVKR ISGLIYEETR GVLKVFLENV IRDAVTYTEH AKRKTVTAMD VVYALKRQGR TLYGFGG UniProtKB: Histone H4 |
-Macromolecule #4: Histone H2A
| Macromolecule | Name: Histone H2A / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 12.660719 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: AKTRSSRAGL QFPVGRVHRL LRKGNYAERV GAGAPVYLAA VLEYLTAEIL ELAGNAARDN KKTRIIPRHL QLAVRNDEEL NKLLGRVTI AQGGVLPNIQ SVLLPKKTES SKSAKSK UniProtKB: Histone H2A |
-Macromolecule #5: Histone H2B 1.1
| Macromolecule | Name: Histone H2B 1.1 / type: protein_or_peptide / ID: 5 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 10.478032 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: TRKESYAIYV YKVLKQVHPD TGISSKAMSI MNSFVNDVFE RIAGEASRLA HYNKRSTITS REIQTAVRLL LPGELAKHAV SEGTKAVTK YTSAK UniProtKB: Histone H2B 1.1 |
-Macromolecule #6: Histone-lysine N-methyltransferase, H3 lysine-79 specific
| Macromolecule | Name: Histone-lysine N-methyltransferase, H3 lysine-79 specific type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO / EC number: histone-lysine N-methyltransferase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.930039 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: LELRLKSPVG AEPAVYPWPL PVYDKHHDAA HEIIETIRWV CEEIPDLKLA MENYVLIDYD TKSFESMQRL CDKYNRAIDS IHQLWKGTT QPMKLNTRPS TGLLRHILQQ VYNHSVTDPE KLNNYEPFSP EVYGETSFDL VAQMIDEIKM TDDDLFVDLG S GVGQVVLQ ...String: LELRLKSPVG AEPAVYPWPL PVYDKHHDAA HEIIETIRWV CEEIPDLKLA MENYVLIDYD TKSFESMQRL CDKYNRAIDS IHQLWKGTT QPMKLNTRPS TGLLRHILQQ VYNHSVTDPE KLNNYEPFSP EVYGETSFDL VAQMIDEIKM TDDDLFVDLG S GVGQVVLQ VAAATNCKHH YGVEKADIPA KYAETMDREF RKWMKWYGKK HAEYTLERGD FLSEEWRERI ANTSVIFVNN FA FGPEVDH QLKERFANMK EGGRIVSSKP FAPLNFRINS RNLSDIGTIM RVVELSPLKG SVSWTGKPVS YYLHTIDRTI LEN YFSSLK NP UniProtKB: Histone-lysine N-methyltransferase, H3 lysine-79 specific |
-Macromolecule #7: Ubiquitin
| Macromolecule | Name: Ubiquitin / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 8.576831 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MQIFVKTLTG KTITLEVEPS DTIENVKAKI QDKEGIPPDQ QRLIFAGKQL EDGRTLSDYN IQKESTLHLV LRLRGG UniProtKB: Polyubiquitin-B |
-Macromolecule #8: S-ADENOSYLMETHIONINE
| Macromolecule | Name: S-ADENOSYLMETHIONINE / type: ligand / ID: 8 / Number of copies: 1 / Formula: SAM |
|---|---|
| Molecular weight | Theoretical: 398.437 Da |
| Chemical component information |  ChemComp-SAM: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 37.28 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-6jma: |
 Movie
Movie Controller
Controller






























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)