[English] 日本語
 Yorodumi
Yorodumi- EMDB-9781: Structure of human voltage-gated sodium channel Nav1.7 in complex... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9781 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of human voltage-gated sodium channel Nav1.7 in complex with auxiliary beta subunits, huwentoxin-IV and saxitoxin | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | voltage-gated sodium channel / MEMBRANE PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationresponse to pyrethroid / corticospinal neuron axon guidance / positive regulation of voltage-gated sodium channel activity / action potential propagation / detection of mechanical stimulus involved in sensory perception / voltage-gated sodium channel activity involved in cardiac muscle cell action potential / regulation of atrial cardiac muscle cell membrane depolarization / voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization / membrane depolarization during Purkinje myocyte cell action potential / cardiac conduction ...response to pyrethroid / corticospinal neuron axon guidance / positive regulation of voltage-gated sodium channel activity / action potential propagation / detection of mechanical stimulus involved in sensory perception / voltage-gated sodium channel activity involved in cardiac muscle cell action potential / regulation of atrial cardiac muscle cell membrane depolarization / voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization / membrane depolarization during Purkinje myocyte cell action potential / cardiac conduction / membrane depolarization during cardiac muscle cell action potential / membrane depolarization during action potential / positive regulation of sodium ion transport / regulation of sodium ion transmembrane transport / axon initial segment / regulation of ventricular cardiac muscle cell membrane repolarization / cardiac muscle cell action potential involved in contraction / node of Ranvier / voltage-gated sodium channel complex / sodium channel inhibitor activity / neuronal action potential propagation / locomotion / Interaction between L1 and Ankyrins / voltage-gated sodium channel activity / detection of temperature stimulus involved in sensory perception of pain / Phase 0 - rapid depolarisation / regulation of heart rate by cardiac conduction / behavioral response to pain / intercalated disc / sodium channel regulator activity / membrane depolarization / neuronal action potential / cardiac muscle contraction / T-tubule / sensory perception of pain / axon terminus / axon guidance / sodium ion transmembrane transport / post-embryonic development / positive regulation of neuron projection development / circadian rhythm / response to toxic substance / Sensory perception of sweet, bitter, and umami (glutamate) taste / nervous system development / response to heat / gene expression / chemical synaptic transmission / perikaryon / transmembrane transporter binding / cell adhesion / inflammatory response / axon / synapse / extracellular region / plasma membrane Similarity search - Function | ||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | ||||||||||||||||||
 Authors Authors | Shen H / Liu D | ||||||||||||||||||
| Funding support |  China, 5 items China, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Science / Year: 2019 Journal: Science / Year: 2019Title: Structures of human Na1.7 channel in complex with auxiliary subunits and animal toxins. Authors: Huaizong Shen / Dongliang Liu / Kun Wu / Jianlin Lei / Nieng Yan /  Abstract: Voltage-gated sodium channel Na1.7 represents a promising target for pain relief. Here we report the cryo-electron microscopy structures of the human Na1.7-β1-β2 complex bound to two combinations ...Voltage-gated sodium channel Na1.7 represents a promising target for pain relief. Here we report the cryo-electron microscopy structures of the human Na1.7-β1-β2 complex bound to two combinations of pore blockers and gating modifier toxins (GMTs), tetrodotoxin with protoxin-II and saxitoxin with huwentoxin-IV, both determined at overall resolutions of 3.2 angstroms. The two structures are nearly identical except for minor shifts of voltage-sensing domain II (VSD), whose S3-S4 linker accommodates the two GMTs in a similar manner. One additional protoxin-II sits on top of the S3-S4 linker in VSD The structures may represent an inactivated state with all four VSDs "up" and the intracellular gate closed. The structures illuminate the path toward mechanistic understanding of the function and disease of Na1.7 and establish the foundation for structure-aided development of analgesics. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9781.map.gz emd_9781.map.gz | 49.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9781-v30.xml emd-9781-v30.xml emd-9781.xml emd-9781.xml | 22.3 KB 22.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9781.png emd_9781.png | 107.8 KB | ||
| Filedesc metadata |  emd-9781.cif.gz emd-9781.cif.gz | 8.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9781 http://ftp.pdbj.org/pub/emdb/structures/EMD-9781 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9781 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9781 | HTTPS FTP |
-Related structure data
| Related structure data |  6j8gMC  6j8hMC  9782C  6j8iC  6j8jC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9781.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9781.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.091 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Complex of human voltage-gated sodium channel Nav1.7 with auxilia...
| Entire | Name: Complex of human voltage-gated sodium channel Nav1.7 with auxiliary beta subunits, huwentoxin-IV and saxitoxin |
|---|---|
| Components |
|
-Supramolecule #1: Complex of human voltage-gated sodium channel Nav1.7 with auxilia...
| Supramolecule | Name: Complex of human voltage-gated sodium channel Nav1.7 with auxiliary beta subunits, huwentoxin-IV and saxitoxin type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Sodium channel subunit beta-2
| Macromolecule | Name: Sodium channel subunit beta-2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.355859 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MHRDAWLPRP AFSLTGLSLF FSLVPPGRSM EVTVPATLNV LNGSDARLPC TFNSCYTVNH KQFSLNWTYQ ECNNCSEEMF LQFRMKIIN LKLERFQDRV EFSGNPSKYD VSVMLRNVQP EDEGIYNCYI MNPPDRHRGH GKIHLQVLME EPPERDSTVA V IVGASVGG ...String: MHRDAWLPRP AFSLTGLSLF FSLVPPGRSM EVTVPATLNV LNGSDARLPC TFNSCYTVNH KQFSLNWTYQ ECNNCSEEMF LQFRMKIIN LKLERFQDRV EFSGNPSKYD VSVMLRNVQP EDEGIYNCYI MNPPDRHRGH GKIHLQVLME EPPERDSTVA V IVGASVGG FLAVVILVLM VVKCVRRKKE QKLSTDDLKT EEEGKTDGEG NPDDGAK UniProtKB: Sodium channel regulatory subunit beta-2 |
-Macromolecule #2: Sodium channel protein type 9 subunit alpha
| Macromolecule | Name: Sodium channel protein type 9 subunit alpha / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 231.211922 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MASWSHPQFE KGGGARGGSG GGSWSHPQFE KGFDYKDDDD KGTMAMLPPP GPQSFVHFTK QSLALIEQRI AERKSKEPKE EKKDDDEEA PKPSSDLEAG KQLPFIYGDI PPGMVSEPLE DLDPYYADKK TFIVLNKGKT IFRFNATPAL YMLSPFSPLR R ISIKILVH ...String: MASWSHPQFE KGGGARGGSG GGSWSHPQFE KGFDYKDDDD KGTMAMLPPP GPQSFVHFTK QSLALIEQRI AERKSKEPKE EKKDDDEEA PKPSSDLEAG KQLPFIYGDI PPGMVSEPLE DLDPYYADKK TFIVLNKGKT IFRFNATPAL YMLSPFSPLR R ISIKILVH SLFSMLIMCT ILTNCIFMTM NNPPDWTKNV EYTFTGIYTF ESLVKILARG FCVGEFTFLR DPWNWLDFVV IV FAYLTEF VNLGNVSALR TFRVLRALKT ISVIPGLKTI VGALIQSVKK LSDVMILTVF CLSVFALIGL QLFMGNLKHK CFR NSLENN ETLESIMNTL ESEEDFRKYF YYLEGSKDAL LCGFSTDSGQ CPEGYTCVKI GRNPDYGYTS FDTFSWAFLA LFRL MTQDY WENLYQQTLR AAGKTYMIFF VVVIFLGSFY LINLILAVVA MAYKEQNQAN IEEAKQKELE FQQMLDRLKK EQEEA EAIA AAAAEYTSIR RSRIMGLSES SSETSKLSSK SAKERRNRRK KKNQKKLSSG EEKGDAEKLS KSESEDSIRR KSFHLG VEG HRRAHEKRLS TPNQSPLSIR GSLFSARRSS RTSLFSFKGR GRDIGSETEF ADDEHSIFGD NESRRGSLFV PHRPQER RS SNISQASRSP PMLPVNGKMH SAVDCNGVVS LVDGRSALML PNGQLLPEVI IDKATSDDSG TTNQIHKKRR CSSYLLSE D MLNDPNLRQR AMSRASILTN TVEELEESRQ KCPPWWYRFA HKFLIWNCSP YWIKFKKCIY FIVMDPFVDL AITICIVLN TLFMAMEHHP MTEEFKNVLA IGNLVFTGIF AAEMVLKLIA MDPYEYFQVG WNIFDSLIVT LSLVELFLAD VEGLSVLRSF RLLRVFKLA KSWPTLNMLI KIIGNSVGAL GNLTLVLAII VFIFAVVGMQ LFGKSYKECV CKINDDCTLP RWHMNDFFHS F LIVFRVLC GEWIETMWDC MEVAGQAMCL IVYMMVMVIG NLVVLNLFLA LLLSSFSSDN LTAIEEDPDA NNLQIAVTRI KK GINYVKQ TLREFILKAF SKKPKISREI RQAEDLNTKK ENYISNHTLA EMSKGHNFLK EKDKISGFGS SVDKHLMEDS DGQ SFIHNP SLTVTVPIAP GESDLENMNA EELSSDSDSE YSKVRLNRSS SSECSTVDNP LPGEGEEAEA EPMNSDEPEA CFTD GCVWR FSCCQVNIES GKGKIWWNIR KTCYKIVEHS WFESFIVLMI LLSSGALAFE DIYIERKKTI KIILEYADKI FTYIF ILEM LLKWIAYGYK TYFTNAWCWL DFLIVDVSLV TLVANTLGYS DLGPIKSLRT LRALRPLRAL SRFEGMRVVV NALIGA IPS IMNVLLVCLI FWLIFSIMGV NLFAGKFYEC INTTDGSRFP ASQVPNRSEC FALMNVSQNV RWKNLKVNFD NVGLGYL SL LQVATFKGWT IIMYAAVDSV NVDKQPKYEY SLYMYIYFVV FIIFGSFFTL NLFIGVIIDN FNQQKKKLGG QDIFMTEE Q KKYYNAMKKL GSKKPQKPIP RPGNKIQGCI FDLVTNQAFD ISIMVLICLN MVTMMVEKEG QSQHMTEVLY WINVVFIIL FTGECVLKLI SLRHYYFTVG WNIFDFVVVI ISIVGMFLAD LIETYFVSPT LFRVIRLARI GRILRLVKGA KGIRTLLFAL MMSLPALFN IGLLLFLVMF IYAIFGMSNF AYVKKEDGIN DMFNFETFGN SMICLFQITT SAGWDGLLAP ILNSKPPDCD P KKVHPGSS VEGDCGNPSV GIFYFVSYII ISFLVVVNMY IAVILENFSV ATEESTEPLS EDDFEMFYEV WEKFDPDATQ FI EFSKLSD FAAALDPPLL IAKPNKVQLI AMDLPMVSGD RIHCLDILFA FTKRVLGESG EMDSLRSQME ERFMSANPSK VSY EPITTT LKRKQEDVSA TVIQRAYRRY RLRQNVKNIS SIYIKDGDRD DDLLNKKDMA FDNVNENSSP EKTDATSSTT SPPS YDSVT KPDKEKYEQD RTEKEDKGKD SKESKK UniProtKB: Sodium channel protein type 9 subunit alpha |
-Macromolecule #3: Sodium channel subunit beta-1
| Macromolecule | Name: Sodium channel subunit beta-1 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.732115 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGRLLALVVG AALVSSACGG CVEVDSETEA VYGMTFKILC ISCKRRSETN AETFTEWTFR QKGTEEFVKI LRYENEVLQL EEDERFEGR VVWNGSRGTK DLQDLSIFIT NVTYNHSGDY ECHVYRLLFF ENYEHNTSVV KKIHIEVVDK ANRDMASIVS E IMMYVLIV ...String: MGRLLALVVG AALVSSACGG CVEVDSETEA VYGMTFKILC ISCKRRSETN AETFTEWTFR QKGTEEFVKI LRYENEVLQL EEDERFEGR VVWNGSRGTK DLQDLSIFIT NVTYNHSGDY ECHVYRLLFF ENYEHNTSVV KKIHIEVVDK ANRDMASIVS E IMMYVLIV VLTIWLVAEM IYCYKKIAAA TETAAQENAS EYLAITSESK ENCTGVQVAE UniProtKB: Sodium channel regulatory subunit beta-1 |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 7 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #6: [(3aS,4R,10aS)-2,6-diamino-10,10-dihydroxy-3a,4,9,10-tetrahydro-3...
| Macromolecule | Name: [(3aS,4R,10aS)-2,6-diamino-10,10-dihydroxy-3a,4,9,10-tetrahydro-3H,8H-pyrrolo[1,2-c]purin-4-yl]methyl carbamate type: ligand / ID: 6 / Number of copies: 1 / Formula: 9SL |
|---|---|
| Molecular weight | Theoretical: 299.286 Da |
| Chemical component information | 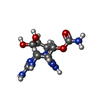 ChemComp-9SL: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















