+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9401 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CH505 SOSIP.664 trimer in complex with DH522.2 Fab | |||||||||
 Map data Map data | CH505 SOSIP.664 trimer in complex with DH522.2 Fab | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   Human immunodeficiency virus 1 Human immunodeficiency virus 1 | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 28.1 Å | |||||||||
 Authors Authors | Fera D / Harrison SC | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2017 Journal: Nat Commun / Year: 2017Title: Initiation of HIV neutralizing B cell lineages with sequential envelope immunizations. Authors: Wilton B Williams / Jinsong Zhang / Chuancang Jiang / Nathan I Nicely / Daniela Fera / Kan Luo / M Anthony Moody / Hua-Xin Liao / S Munir Alam / Thomas B Kepler / Akshaya Ramesh / Kevin ...Authors: Wilton B Williams / Jinsong Zhang / Chuancang Jiang / Nathan I Nicely / Daniela Fera / Kan Luo / M Anthony Moody / Hua-Xin Liao / S Munir Alam / Thomas B Kepler / Akshaya Ramesh / Kevin Wiehe / James A Holland / Todd Bradley / Nathan Vandergrift / Kevin O Saunders / Robert Parks / Andrew Foulger / Shi-Mao Xia / Mattia Bonsignori / David C Montefiori / Mark Louder / Amanda Eaton / Sampa Santra / Richard Scearce / Laura Sutherland / Amanda Newman / Hilary Bouton-Verville / Cindy Bowman / Howard Bomze / Feng Gao / Dawn J Marshall / John F Whitesides / Xiaoyan Nie / Garnett Kelsoe / Steven G Reed / Christopher B Fox / Kim Clary / Marguerite Koutsoukos / David Franco / John R Mascola / Stephen C Harrison / Barton F Haynes / Laurent Verkoczy /    Abstract: A strategy for HIV-1 vaccine development is to define envelope (Env) evolution of broadly neutralizing antibodies (bnAbs) in infection and to recreate those events by vaccination. Here, we report ...A strategy for HIV-1 vaccine development is to define envelope (Env) evolution of broadly neutralizing antibodies (bnAbs) in infection and to recreate those events by vaccination. Here, we report host tolerance mechanisms that limit the development of CD4-binding site (CD4bs), HCDR3-binder bnAbs via sequential HIV-1 Env vaccination. Vaccine-induced macaque CD4bs antibodies neutralize 7% of HIV-1 strains, recognize open Env trimers, and accumulate relatively modest somatic mutations. In naive CD4bs, unmutated common ancestor knock-in mice EnvB cell clones develop anergy and partial deletion at the transitional to mature B cell stage, but become Env upon receptor editing. In comparison with repetitive Env immunizations, sequential Env administration rescue anergic Env (non-edited) precursor B cells. Thus, stepwise immunization initiates CD4bs-bnAb responses, but immune tolerance mechanisms restrict their development, suggesting that sequential immunogen-based vaccine regimens will likely need to incorporate strategies to expand bnAb precursor pools. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9401.map.gz emd_9401.map.gz | 2.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9401-v30.xml emd-9401-v30.xml emd-9401.xml emd-9401.xml | 13.9 KB 13.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9401.png emd_9401.png | 22.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9401 http://ftp.pdbj.org/pub/emdb/structures/EMD-9401 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9401 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9401 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9401.map.gz / Format: CCP4 / Size: 18.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9401.map.gz / Format: CCP4 / Size: 18.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CH505 SOSIP.664 trimer in complex with DH522.2 Fab | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.13 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
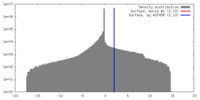
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : CH505 SOSIP.664 in complex with DH522.2 Fab
| Entire | Name: CH505 SOSIP.664 in complex with DH522.2 Fab |
|---|---|
| Components |
|
-Supramolecule #1: CH505 SOSIP.664 in complex with DH522.2 Fab
| Supramolecule | Name: CH505 SOSIP.664 in complex with DH522.2 Fab / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: CH505 SOSIP.664
| Supramolecule | Name: CH505 SOSIP.664 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: DH522.2 Fab
| Supramolecule | Name: DH522.2 Fab / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: CH505 SOSIP.664
| Macromolecule | Name: CH505 SOSIP.664 / type: other / ID: 1 / Classification: other |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Sequence | String: AENLWVTVYY GVPVWKEAKT TLFCASDAKA YEKEVHNVWA THACVPTDPN PQEMVLKNVT ENFNMWKNDM VDQMHEDVIS LWDQSLKPCV KLTPLCVTLN CTNATASNSS IIEGMKNCSF NITTELRDKR EKKNALFYKL DIVQLDGNSS QYRLINCNTS VITQACPKVS ...String: AENLWVTVYY GVPVWKEAKT TLFCASDAKA YEKEVHNVWA THACVPTDPN PQEMVLKNVT ENFNMWKNDM VDQMHEDVIS LWDQSLKPCV KLTPLCVTLN CTNATASNSS IIEGMKNCSF NITTELRDKR EKKNALFYKL DIVQLDGNSS QYRLINCNTS VITQACPKVS FDPIPIHYCA PAGYAILKCN NKTFTGTGPC NNVSTVQCTH GIKPVVSTQL LLNGSLAEGE IIIRSENITN NVKTIIVHLN ESVKIECTRP NNKTRTSIRI GPGQAFYATG QVIGDIREAY CNINESKWNE TLQRVSKKLK EYFPHKNITF QPSSGGDLEI TTHSFNCGGE FFYCNTSSLF NRTYMANSTD MANSTETNST RTITIHCRIK QIINMWQEVG RAMYAPPIAG NITCISNITG LLLTRDGGKN NTETFRPGGG NMKDNWRSEL YKYKVVKIEP LGVAPTRCKR RVVGRRRRRR AVGIGAVFLG FLGAAGSTMG AASMTLTVQA RNLLSGIVQQ QSNLLRAPEA QQHLLKLTVW GIKQLQARVL AVERYLRDQQ LLGIWGCSGK LICCTNVPWN SSWSNRNLSE IWDNMTWLQW DKEISNYTQI IYGLLEESQN QQEKNEQDLL ALD |
-Macromolecule #2: DH522.2 Fab Heavy Chain
| Macromolecule | Name: DH522.2 Fab Heavy Chain / type: other / ID: 2 / Classification: other |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLQESGPG LVKPSETLSL TCAVSGGSIG DDYYWNWIRQ SPGKGLEWIG SIYGSFGGTN FNPSLKNRVT ISMDTSNNQV SLKLNSVTAA DTAVYYCARG SHSIVVLFGY YFDYWGQGVL VTVSSASTKG PSVFPLAPSS RSTSESTAAL GCLVKD YFP EPVTVSWNSG ...String: QVQLQESGPG LVKPSETLSL TCAVSGGSIG DDYYWNWIRQ SPGKGLEWIG SIYGSFGGTN FNPSLKNRVT ISMDTSNNQV SLKLNSVTAA DTAVYYCARG SHSIVVLFGY YFDYWGQGVL VTVSSASTKG PSVFPLAPSS RSTSESTAAL GCLVKD YFP EPVTVSWNSG SLTSGVHTFP AVLQSSGLYS LSSVVTVPSS SLGTQTYVCN VNHKPSN TK VDKRVEIKT |
-Macromolecule #3: DH522.2 Fab Light Chain
| Macromolecule | Name: DH522.2 Fab Light Chain / type: other / ID: 3 / Classification: other |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QSALTQPPSV SKSLGQSVTI SCSGTTNDIG AYNGVS WYQ HHSDTAPRLL IYEVNKRPSG VSDRFSGSKS GNTASLTISG LQAEDEADYY CGSYRSG ST WVFGGGTRLT VLGQPKASPT VTLFPPSSEE LQANKATLVC LISDFYPGVV KVAWKADG S AVNAGVETTT ...String: QSALTQPPSV SKSLGQSVTI SCSGTTNDIG AYNGVS WYQ HHSDTAPRLL IYEVNKRPSG VSDRFSGSKS GNTASLTISG LQAEDEADYY CGSYRSG ST WVFGGGTRLT VLGQPKASPT VTLFPPSSEE LQANKATLVC LISDFYPGVV KVAWKADG S AVNAGVETTT PSKQSNNKYA ASSYLSLTSD QWKSHKSYSC QVTHEGSTVE KTVAPAECS |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.01 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Staining | Type: NEGATIVE / Material: uranyl formate |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Image recording | Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 28.1 Å / Resolution method: FSC 0.5 CUT-OFF / Number images used: 15269 |
|---|---|
| Initial angle assignment | Type: COMMON LINE |
| Final angle assignment | Type: COMMON LINE |
 Movie
Movie Controller
Controller



 UCSF Chimera
UCSF Chimera






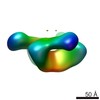



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)