+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9139 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | H7 HA0 in complex with Fab from H7.5 IgG | |||||||||
 Map data Map data | H7 HA0 in complex with Fv from H7.5 IgG | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Hemagglutinin / antibody / Fab / H7 / HA0 / protein complex / VIRAL PROTEIN / VIRAL PROTEIN-Immune System complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationviral budding from plasma membrane / clathrin-dependent endocytosis of virus by host cell / host cell surface receptor binding / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / membrane Similarity search - Function | |||||||||
| Biological species |  Influenza A virus (A/New York/107/2003(H7N2)) / Influenza A virus (A/New York/107/2003(H7N2)) /   Influenza A virus / Influenza A virus /  Homo sapiens (human) Homo sapiens (human) | |||||||||
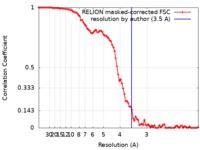
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Pallesen J / Turner HL | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: PLoS Biol / Year: 2019 Journal: PLoS Biol / Year: 2019Title: Potent anti-influenza H7 human monoclonal antibody induces separation of hemagglutinin receptor-binding head domains. Authors: Hannah L Turner / Jesper Pallesen / Shanshan Lang / Sandhya Bangaru / Sarah Urata / Sheng Li / Christopher A Cottrell / Charles A Bowman / James E Crowe / Ian A Wilson / Andrew B Ward /  Abstract: Seasonal influenza virus infections can cause significant morbidity and mortality, but the threat from the emergence of a new pandemic influenza strain might have potentially even more devastating ...Seasonal influenza virus infections can cause significant morbidity and mortality, but the threat from the emergence of a new pandemic influenza strain might have potentially even more devastating consequences. As such, there is intense interest in isolating and characterizing potent neutralizing antibodies that target the hemagglutinin (HA) viral surface glycoprotein. Here, we use cryo-electron microscopy (cryoEM) to decipher the mechanism of action of a potent HA head-directed monoclonal antibody (mAb) bound to an influenza H7 HA. The epitope of the antibody is not solvent accessible in the compact, prefusion conformation that typifies all HA structures to date. Instead, the antibody binds between HA head protomers to an epitope that must be partly or transiently exposed in the prefusion conformation. The "breathing" of the HA protomers is implied by the exposure of this epitope, which is consistent with metastability of class I fusion proteins. This structure likely therefore represents an early structural intermediate in the viral fusion process. Understanding the extent of transient exposure of conserved neutralizing epitopes also may lead to new opportunities to combat influenza that have not been appreciated previously. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9139.map.gz emd_9139.map.gz | 59.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9139-v30.xml emd-9139-v30.xml emd-9139.xml emd-9139.xml | 22.8 KB 22.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9139_fsc.xml emd_9139_fsc.xml | 9 KB | Display |  FSC data file FSC data file |
| Images |  emd_9139.png emd_9139.png | 69.6 KB | ||
| Filedesc metadata |  emd-9139.cif.gz emd-9139.cif.gz | 6.9 KB | ||
| Others |  emd_9139_additional.map.gz emd_9139_additional.map.gz emd_9139_half_map_1.map.gz emd_9139_half_map_1.map.gz emd_9139_half_map_2.map.gz emd_9139_half_map_2.map.gz | 58.4 MB 49.7 MB 49.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9139 http://ftp.pdbj.org/pub/emdb/structures/EMD-9139 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9139 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9139 | HTTPS FTP |
-Validation report
| Summary document |  emd_9139_validation.pdf.gz emd_9139_validation.pdf.gz | 940.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_9139_full_validation.pdf.gz emd_9139_full_validation.pdf.gz | 940.4 KB | Display | |
| Data in XML |  emd_9139_validation.xml.gz emd_9139_validation.xml.gz | 16.1 KB | Display | |
| Data in CIF |  emd_9139_validation.cif.gz emd_9139_validation.cif.gz | 21.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9139 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9139 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9139 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9139 | HTTPS FTP |
-Related structure data
| Related structure data |  6mlmMC  9142C  9143C  9144C  9145C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9139.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9139.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | H7 HA0 in complex with Fv from H7.5 IgG | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.02 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
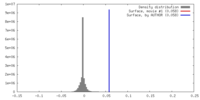
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: H7 HA0 in complex with Fv from H7.5 IgG
| File | emd_9139_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | H7 HA0 in complex with Fv from H7.5 IgG | ||||||||||||
| Projections & Slices |
| ||||||||||||
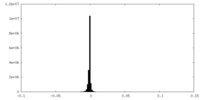
| Density Histograms |
-Half map: H7 HA0 in complex with Fv from H7.5 IgG
| File | emd_9139_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | H7 HA0 in complex with Fv from H7.5 IgG | ||||||||||||
| Projections & Slices |
| ||||||||||||
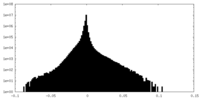
| Density Histograms |
-Half map: H7 HA0 in complex with Fv from H7.5 IgG
| File | emd_9139_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | H7 HA0 in complex with Fv from H7.5 IgG | ||||||||||||
| Projections & Slices |
| ||||||||||||
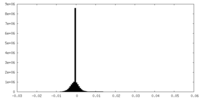
| Density Histograms |
- Sample components
Sample components
-Entire : H7 HA0 in complex with H7.5 Fab
| Entire | Name: H7 HA0 in complex with H7.5 Fab |
|---|---|
| Components |
|
-Supramolecule #1: H7 HA0 in complex with H7.5 Fab
| Supramolecule | Name: H7 HA0 in complex with H7.5 Fab / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Influenza A virus (A/New York/107/2003(H7N2)) Influenza A virus (A/New York/107/2003(H7N2)) |
| Molecular weight | Theoretical: 230 KDa |
-Macromolecule #1: Hemagglutinin HA1 chain
| Macromolecule | Name: Hemagglutinin HA1 chain / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Influenza A virus (A/New York/107/2003(H7N2)) / Strain: A/New York/107/2003(H7N2) Influenza A virus (A/New York/107/2003(H7N2)) / Strain: A/New York/107/2003(H7N2) |
| Molecular weight | Theoretical: 36.01882 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MNTQILAFIA CVLTGVKGDK ICLGHHAVAN GTKVNTLTER GIEVVNATET VETTNIKKIC TQGKRPTDLG QCGLLGTLIG PPQCDQFLE FSSDLIIERR EGTDICYPGR FTNEESLRQI LRRSGGIGKE SMGFTYSGIR TNGATSACTR SGSSFYAEMK W LLSNSDNA ...String: MNTQILAFIA CVLTGVKGDK ICLGHHAVAN GTKVNTLTER GIEVVNATET VETTNIKKIC TQGKRPTDLG QCGLLGTLIG PPQCDQFLE FSSDLIIERR EGTDICYPGR FTNEESLRQI LRRSGGIGKE SMGFTYSGIR TNGATSACTR SGSSFYAEMK W LLSNSDNA AFPQMTKAYR NPRNKPALII WGVHHSESVS EQTKLYGSGN KLITVRSSKY QQSFTPNPGA RRIDFHWLLL DP NDTVTFT FNGAFIAPDR TSFFRGESLG VQSDAPLDSS CRGDCFHSGG TIVSSLPFQN INSRTVGKCP RYVKQKSLLL ATG MRNVPE KP UniProtKB: Hemagglutinin |
-Macromolecule #2: Hemagglutinin HA2 chain
| Macromolecule | Name: Hemagglutinin HA2 chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Influenza A virus / Strain: A/New York/107/2003(H7N2) Influenza A virus / Strain: A/New York/107/2003(H7N2) |
| Molecular weight | Theoretical: 25.583803 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: KPRGLFGAIA GFIENGWEGL INGWYGFRHQ NAQGEGTAAD YKSTQSAIDQ ITGKLNRLIG KTNQQFELID NEFNEIEQQI GNVINWTRD AMTEIWSYNA ELLVAMENQH TIDLADSEMS KLYERVKKQL RENAEEDGTG CFEIFHKCDD QCMESIRNNT Y DHTQYRTE ...String: KPRGLFGAIA GFIENGWEGL INGWYGFRHQ NAQGEGTAAD YKSTQSAIDQ ITGKLNRLIG KTNQQFELID NEFNEIEQQI GNVINWTRD AMTEIWSYNA ELLVAMENQH TIDLADSEMS KLYERVKKQL RENAEEDGTG CFEIFHKCDD QCMESIRNNT Y DHTQYRTE SLQNRIQIDP VKLSSGYKDI ILWFSFGASC FLLLAIAMGL VFICIKNGNM QCTICI UniProtKB: Hemagglutinin |
-Macromolecule #3: Heavy chain Fv of H7.5 Fab
| Macromolecule | Name: Heavy chain Fv of H7.5 Fab / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.441402 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: VQLVQSGAEV KKPGASVKVS CKASGYTLTR YYFHWVRQAP GQGFEWMGII NPNGGGTSYA QKFGDRVIMT SDMSTSTIYM ELSSLRSED TAVYYCARDM PYYHDSGGPL FDLWGQGTLV TVSSASTKGP SVFPLAPSGG TAALGCLVKD YFPEPVTVSW N SGALTSGV ...String: VQLVQSGAEV KKPGASVKVS CKASGYTLTR YYFHWVRQAP GQGFEWMGII NPNGGGTSYA QKFGDRVIMT SDMSTSTIYM ELSSLRSED TAVYYCARDM PYYHDSGGPL FDLWGQGTLV TVSSASTKGP SVFPLAPSGG TAALGCLVKD YFPEPVTVSW N SGALTSGV HTFPAVLQSS GLYSLSSVVT VPSSSLGTQT YICNVNHKPS NTKVDKKVEP K |
-Macromolecule #4: Light chain Fv of H7.5 Fab
| Macromolecule | Name: Light chain Fv of H7.5 Fab / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.340836 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EIVMTQSPSS LSASVGDRVT ITCRPSQSIS TFLNWYEQKP GKAPKLLIYD ASSLQSGVPS RFSGSGSGTE FTLTISSLQP EDFATYYCQ QSFSTPYTFG QGTRLEIKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS ...String: EIVMTQSPSS LSASVGDRVT ITCRPSQSIS TFLNWYEQKP GKAPKLLIYD ASSLQSGVPS RFSGSGSGTE FTLTISSLQP EDFATYYCQ QSFSTPYTFG QGTRLEIKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS KDSTYSLSST LTLSKADYEK HKVYACEVTH QGLSSPVTKS FNRGE |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 9 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average exposure time: 7.0 sec. / Average electron dose: 67.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)