[English] 日本語
 Yorodumi
Yorodumi- EMDB-9006: Trans-envelope channel formed during initial infection of bacteri... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9006 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
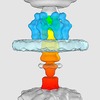
| Title | Trans-envelope channel formed during initial infection of bacteriophage P22 in Salmonella | |||||||||
 Map data Map data | Trans-envelope channel formed during initial infection of bacteriophage P22 in Salmonella | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Enterobacteria phage P22 (virus) Enterobacteria phage P22 (virus) | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 28.0 Å | |||||||||
 Authors Authors | Liu J / Molineux IJ / Wang CY | |||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2019 Journal: Nat Microbiol / Year: 2019Title: Structural dynamics of bacteriophage P22 infection initiation revealed by cryo-electron tomography. Authors: Chunyan Wang / Jiagang Tu / Jun Liu / Ian J Molineux /  Abstract: For successful infection, bacteriophages must overcome multiple barriers to transport their genome and proteins across the bacterial cell envelope. We use cryo-electron tomography to study the ...For successful infection, bacteriophages must overcome multiple barriers to transport their genome and proteins across the bacterial cell envelope. We use cryo-electron tomography to study the infection initiation of phage P22 in Salmonella enterica serovar Typhimurium, revealing how a channel forms to allow genome translocation into the cytoplasm. Our results show free phages that initially attach obliquely to the cell through interactions between the O antigen and two of the six tailspikes; the tail needle also abuts the cell surface. The virion then orients perpendicularly and the needle penetrates the outer membrane. The needle is released and the internal head protein gp7* is ejected and assembles into an extracellular channel that extends from the gp10 baseplate to the cell surface. A second protein, gp20, is ejected and assembles into a structure that extends the extracellular channel across the outer membrane into the periplasm. Insertion of the third ejected protein, gp16, into the cytoplasmic membrane probably completes the overall trans-envelope channel into the cytoplasm. Construction of a trans-envelope channel is an essential step during infection of Gram-negative bacteria by all short-tailed phages, because such virions cannot directly deliver their genome into the cell cytoplasm. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9006.map.gz emd_9006.map.gz | 3.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9006-v30.xml emd-9006-v30.xml emd-9006.xml emd-9006.xml | 7.6 KB 7.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9006.png emd_9006.png | 110.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9006 http://ftp.pdbj.org/pub/emdb/structures/EMD-9006 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9006 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9006 | HTTPS FTP |
-Validation report
| Summary document |  emd_9006_validation.pdf.gz emd_9006_validation.pdf.gz | 78.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_9006_full_validation.pdf.gz emd_9006_full_validation.pdf.gz | 77.8 KB | Display | |
| Data in XML |  emd_9006_validation.xml.gz emd_9006_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9006 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9006 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9006 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9006 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9006.map.gz / Format: CCP4 / Size: 6.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9006.map.gz / Format: CCP4 / Size: 6.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Trans-envelope channel formed during initial infection of bacteriophage P22 in Salmonella | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 5.4 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Channel structure formed by bacteriophage P22 in Salmonella
| Entire | Name: Channel structure formed by bacteriophage P22 in Salmonella |
|---|---|
| Components |
|
-Supramolecule #1: Channel structure formed by bacteriophage P22 in Salmonella
| Supramolecule | Name: Channel structure formed by bacteriophage P22 in Salmonella type: organelle_or_cellular_component / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage P22 (virus) Enterobacteria phage P22 (virus) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 5 |
|---|---|
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 2.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C6 (6 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 28.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number subtomograms used: 2250 |
|---|---|
| Extraction | Number tomograms: 80 / Number images used: 7217 |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















