+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8952 | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
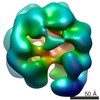
| Title | PTEX Core Complex in the Resetting (Compact) State | ||||||||||||||||||||||||||||||||||||
 Map data Map data | PTEX Core Complex in the Resetting (Compact) State | ||||||||||||||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||||||||||||||
 Keywords Keywords | Translocon / Membrane Protein / ATPase / PROTEIN TRANSPORT | ||||||||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationPTEX complex / apical complex / symbiont-containing vacuole / translocation of peptides or proteins into host cell cytoplasm / symbiont-containing vacuole membrane / response to unfolded protein / cellular response to heat / response to heat / ATP hydrolysis activity / ATP binding / cytoplasm Similarity search - Function | ||||||||||||||||||||||||||||||||||||
| Biological species |   | ||||||||||||||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.23 Å | ||||||||||||||||||||||||||||||||||||
 Authors Authors | Ho C / Lai M | ||||||||||||||||||||||||||||||||||||
| Funding support |  United States, 11 items United States, 11 items
| ||||||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2018 Journal: Nature / Year: 2018Title: Malaria parasite translocon structure and mechanism of effector export. Authors: Chi-Min Ho / Josh R Beck / Mason Lai / Yanxiang Cui / Daniel E Goldberg / Pascal F Egea / Z Hong Zhou /  Abstract: The putative Plasmodium translocon of exported proteins (PTEX) is essential for transport of malarial effector proteins across a parasite-encasing vacuolar membrane into host erythrocytes, but the ...The putative Plasmodium translocon of exported proteins (PTEX) is essential for transport of malarial effector proteins across a parasite-encasing vacuolar membrane into host erythrocytes, but the mechanism of this process remains unknown. Here we show that PTEX is a bona fide translocon by determining structures of the PTEX core complex at near-atomic resolution using cryo-electron microscopy. We isolated the endogenous PTEX core complex containing EXP2, PTEX150 and HSP101 from Plasmodium falciparum in the 'engaged' and 'resetting' states of endogenous cargo translocation using epitope tags inserted using the CRISPR-Cas9 system. In the structures, EXP2 and PTEX150 interdigitate to form a static, funnel-shaped pseudo-seven-fold-symmetric protein-conducting channel spanning the vacuolar membrane. The spiral-shaped AAA+ HSP101 hexamer is tethered above this funnel, and undergoes pronounced compaction that allows three of six tyrosine-bearing pore loops lining the HSP101 channel to dissociate from the cargo, resetting the translocon for the next threading cycle. Our work reveals the mechanism of P. falciparum effector export, and will inform structure-based design of drugs targeting this unique translocon. | ||||||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8952.map.gz emd_8952.map.gz | 27.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8952-v30.xml emd-8952-v30.xml emd-8952.xml emd-8952.xml | 24 KB 24 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8952.png emd_8952.png | 175.3 KB | ||
| Filedesc metadata |  emd-8952.cif.gz emd-8952.cif.gz | 7.5 KB | ||
| Others |  emd_8952_additional.map.gz emd_8952_additional.map.gz | 23 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8952 http://ftp.pdbj.org/pub/emdb/structures/EMD-8952 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8952 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8952 | HTTPS FTP |
-Related structure data
| Related structure data |  6e11MC  8951C  6e10C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8952.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8952.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | PTEX Core Complex in the Resetting (Compact) State | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: PTEX Core Complex in the Resetting (Compact) State, additional map #1
| File | emd_8952_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | PTEX Core Complex in the Resetting (Compact) State, additional map #1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Plasmodium Translocon of Exported Proteins (PTEX) Core Complex
| Entire | Name: Plasmodium Translocon of Exported Proteins (PTEX) Core Complex |
|---|---|
| Components |
|
-Supramolecule #1: Plasmodium Translocon of Exported Proteins (PTEX) Core Complex
| Supramolecule | Name: Plasmodium Translocon of Exported Proteins (PTEX) Core Complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Unknown (Claw)
| Macromolecule | Name: Unknown (Claw) / type: protein_or_peptide / ID: 1 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 5.124308 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) |
-Macromolecule #2: Heat shock protein 101
| Macromolecule | Name: Heat shock protein 101 / type: protein_or_peptide / ID: 2 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 103.031914 KDa |
| Sequence | String: MTRRYLKYYI FVTLLFFVQV INNVLCAPDN KQEQGKYLNR TINILNAGKN IAKSYGHNKL KPIHILSALA KSDYGSTLFK ENNVNAANL KEYIDIALEQ TRAGAPLDNK SKIVNSAEVK ETLALAEAAA NKYKSPKVDV EHLLSGLSND ELVNEIFNEV Y LTDEAIKA ...String: MTRRYLKYYI FVTLLFFVQV INNVLCAPDN KQEQGKYLNR TINILNAGKN IAKSYGHNKL KPIHILSALA KSDYGSTLFK ENNVNAANL KEYIDIALEQ TRAGAPLDNK SKIVNSAEVK ETLALAEAAA NKYKSPKVDV EHLLSGLSND ELVNEIFNEV Y LTDEAIKA ILKRKFEKTK KDKDGKTGTL YIEQFGSNMN EKVRNGKLQG IYGRDEEIRA IIESLLRYNK NSPVLVGNPG TG KTTIVEG LVYRIEKGDV PKELQGYTVI SLNFRKFTSG TSYRGEFETR MKNIIKELKN KKNKIILFVD EIHLLLGAGK AEG GTDAAN LLKPVLSKGE IKLIGATTIA EYRKFIESCS AFERRFEKIL VEPPSVDMTV KILRSLKSKY ENFYGINITD KALV AAAKI SDRFIKDRYL PDKAIDLLNK ACSFLQVQLS GKPRIIDVTE RDIERLSYEI STLEKDVDKV SKKKYNKLIK EFEEK KEQL KKYYEEYVIT GERLKRKKEI EKKLNDLKEL TQNYVYSNKE PPIELQNSLK EAQQKYLELY KETVAYVEAK THNAMN VDA VYQEHVSYIY LRDSGMPLGS LSFESSKGAL KLYNSLSKSI IGNEDIIKSL SDAVVKAATG MKDPEKPIGT FLFLGPT GV GKTELAKTLA IELFNSKDNL IRVNMSEFTE AHSVSKITGS PPGYVGFSDS GQLTEAVREK PHSVVLFDEL EKAHADVF K VLLQILGDGY INDNHRRNID FSNTIIIMTS NLGAELFKKK LFFDADNSGT PEYKRVMEDV RLSLIKKCKK VFKPEFVNR IDKIGVFEPL NKKNLHKIVA LRFKKLEKRL EEKNIQVSVS EKAIDYIIDQ SYDPELGARP TLIFIESVIM TKFAIMYLKK ELVDDMDVF VDYNSKAKNL VINLSKT UniProtKB: Heat shock protein 101 |
-Macromolecule #3: Exported protein 2
| Macromolecule | Name: Exported protein 2 / type: protein_or_peptide / ID: 3 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 33.458707 KDa |
| Sequence | String: MKVSYIFSFF LLFFVYKNTN TVVCDNGYGD LAATSALTTV IKDPISLTIK DIYEHGVKNP FTKIIHKLKK FIRYRKVLRW SRMWWVLLV REIVGDNTIE KKTEKALREI WDQCTIAVYN NTLNAVESKP LLFLHGILNE CRNNFATKLR QDPSLIVAKI D QIIKSQIY ...String: MKVSYIFSFF LLFFVYKNTN TVVCDNGYGD LAATSALTTV IKDPISLTIK DIYEHGVKNP FTKIIHKLKK FIRYRKVLRW SRMWWVLLV REIVGDNTIE KKTEKALREI WDQCTIAVYN NTLNAVESKP LLFLHGILNE CRNNFATKLR QDPSLIVAKI D QIIKSQIY RFWVSEPYLK IGRSHTLYTH ITPDAVPQLP KECTLKHLSS YMEEKLKSME SKKNIESGKY EFDVDSSETD ST KDDGKPD DDDDDDDNFD DDDNFDDDTV EEEDASGDLF KNEKKDENKE UniProtKB: Exported protein 2 |
-Macromolecule #4: Endogenous cargo polypeptide
| Macromolecule | Name: Endogenous cargo polypeptide / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 528.644 Da |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
-Macromolecule #5: Translocon component PTEX150
| Macromolecule | Name: Translocon component PTEX150 / type: protein_or_peptide / ID: 5 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 112.523328 KDa |
| Sequence | String: MRIIILALLI VCTIINYYCA VQNNGNKSLN VMPTCSMPGN DSDSNDNETG DVDNDKNNEL GNANDNNEMN NENAESKNMQ GENSNNQEQ LNENVHANDD AMYEGTPSSD NPPQENVDAN NNEQEYGPPQ EEPVSENNVE NVEVATDDSG NDNINNNDNF N NNDNYNDN ...String: MRIIILALLI VCTIINYYCA VQNNGNKSLN VMPTCSMPGN DSDSNDNETG DVDNDKNNEL GNANDNNEMN NENAESKNMQ GENSNNQEQ LNENVHANDD AMYEGTPSSD NPPQENVDAN NNEQEYGPPQ EEPVSENNVE NVEVATDDSG NDNINNNDNF N NNDNYNDN DNFNEEPPSD DGNKNEDELT EGNQSDDKPM NEEEATINEM GKITNPFEDM LKGKVDDMDI GKMMNKDNLQ SF LSSLTGN KDGSGKNPLS DMMNIFGVPQ TGKEGAEGGV NKENQMKQIN ELKDKLETML KGAGVNVDKI KDSIKNNDLL KNK QLLKEA ISKLTLDPSM MNMLNNKDGA NGKPFDINPD SMMKMFNALS NENGNLDDLK MKPTDGSFDS FNDGVDNNLV PSNP KGQNN NEEDDEEGGD DDDYDDKSFV VNSKYADNSF EDKFNTFDEK DDDVKYELFG ENEEAEELNN NTTTASSKGD ANNSV NTQE GEGEEESFSA NEENINNNNN HNNKNYNNYN TSQQEEDDNS FNENDEPLIS SSQFDNNKKN KMSVSTHNKK SKNLMD SLD LESTNYGSNS SSSMSNNYNS KNKNSKKNNK KKSSQKDYIR TDGKVSFDMA TLQKTIKNFG GADNEIVQNI LKKYVTI DN DDDNDADEDE DEDDDDDDDL DEDEFSVKDI KKLIEEGILD YEDLTENELR KLAKPDDNFY ELSPYASDEK DLSLNETS G LTNEQLKNFL GQNGTYHMSY DSKSIDYAKQ KKSEKKEDQQ EDDDGFYDAY KQIKNSYDGI PNNFNHEAPQ LIGNNYVFT SIYDTKENLI KFLKKNSEYD LYDDDDKEGG NFKSPLYDKY GGKLQKFKRQ RAFNILKQWR AKEKKLKEKK KKEEMEENKE FDFSKNYNF SSKNDGGVTM FSKDQLEDMV KNFGGKPSAH VTDSFSRKEN PFVPTNTKNN SNDDDDMDNG YVTFDGKNKV S ENDDDEKG NNNDDENDND DSNDEEELDE EEDDN UniProtKB: Translocon component PTEX150 |
-Macromolecule #6: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER / type: ligand / ID: 6 / Number of copies: 12 / Formula: AGS |
|---|---|
| Molecular weight | Theoretical: 523.247 Da |
| Chemical component information |  ChemComp-AGS: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Support film - Material: CARBON / Support film - topology: LACEY / Pretreatment - Type: GLOW DISCHARGE / Details: unspecified |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
| Details | PTEX core complex purified from P. falciparum parasites cultured in human erythrocytes |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Lower energy threshold: -10 eV / Energy filter - Upper energy threshold: 10 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Digitization - Frames/image: 2-50 / Average exposure time: 10.0 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 4.0 µm / Calibrated defocus min: 1.5 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-6e11: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)