+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8839 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | The Hsp90 Co-chaperon R2TP Forms a Hexameric Platform | ||||||||||||||||||
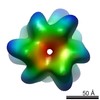
 Map data Map data | Rvb1 and Rvb2 complexed with Tah1 and Pih1. Map with bfactor compensation applied. | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
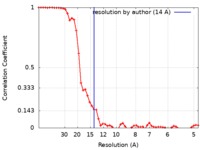
| Method | single particle reconstruction / cryo EM / Resolution: 14.0 Å | ||||||||||||||||||
 Authors Authors | Tian S / Yu G / He H / Liu P / Marshall A / Demeler B / Stagg S / Li H | ||||||||||||||||||
| Funding support |  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Structure / Year: 2017 Journal: Structure / Year: 2017Title: Pih1p-Tah1p Puts a Lid on Hexameric AAA+ ATPases Rvb1/2p. Authors: Shaoxiong Tian / Ge Yu / Huan He / Yu Zhao / Peilu Liu / Alan G Marshall / Borries Demeler / Scott M Stagg / Hong Li /  Abstract: The Saccharomyces cerevisiae (Sc) R2TP complex affords an Hsp90-mediated and nucleotide-driven chaperone activity to proteins of small ribonucleoprotein particles (snoRNPs). The current lack of ...The Saccharomyces cerevisiae (Sc) R2TP complex affords an Hsp90-mediated and nucleotide-driven chaperone activity to proteins of small ribonucleoprotein particles (snoRNPs). The current lack of structural information on the ScR2TP complex, however, prevents a mechanistic understanding of this biological process. We characterized the structure of the ScR2TP complex made up of two AAA+ ATPases, Rvb1/2p, and two Hsp90 binding proteins, Tah1p and Pih1p, and its interaction with the snoRNP protein Nop58p by a combination of analytical ultracentrifugation, isothermal titration calorimetry, chemical crosslinking, hydrogen-deuterium exchange, and cryoelectron microscopy methods. We find that Pih1p-Tah1p interacts with Rvb1/2p cooperatively through the nucleotide-sensitive domain of Rvb1/2p. Nop58p further binds Pih1p-Tahp1 on top of the dome-shaped R2TP. Consequently, nucleotide binding releases Pih1p-Tah1p from Rvb1/2p, which offers a mechanism for nucleotide-driven binding and release of snoRNP intermediates. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8839.map.gz emd_8839.map.gz | 6.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8839-v30.xml emd-8839-v30.xml emd-8839.xml emd-8839.xml | 22.4 KB 22.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8839_fsc.xml emd_8839_fsc.xml | 6.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_8839.png emd_8839.png | 122.1 KB | ||
| Others |  emd_8839_half_map_1.map.gz emd_8839_half_map_1.map.gz emd_8839_half_map_2.map.gz emd_8839_half_map_2.map.gz | 874.4 KB 3.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8839 http://ftp.pdbj.org/pub/emdb/structures/EMD-8839 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8839 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8839 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8839.map.gz / Format: CCP4 / Size: 11.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8839.map.gz / Format: CCP4 / Size: 11.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Rvb1 and Rvb2 complexed with Tah1 and Pih1. Map with bfactor compensation applied. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.43 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
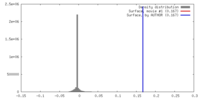
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
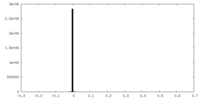
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Rvb1 and Rvb2 complexed with Tah1 and Pih1. Halfmap1
| File | emd_8839_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Rvb1 and Rvb2 complexed with Tah1 and Pih1. Halfmap1 | ||||||||||||
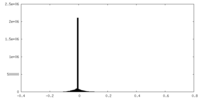
| Projections & Slices |
| ||||||||||||
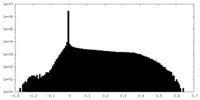
| Density Histograms |
-Half map: Rvb1 and Rvb2 complexed with Tah1 and Pih1. Halfmap2
| File | emd_8839_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Rvb1 and Rvb2 complexed with Tah1 and Pih1. Halfmap2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of Rvb1 and Rvb2 together with Tah1 and Pih1
| Entire | Name: Complex of Rvb1 and Rvb2 together with Tah1 and Pih1 |
|---|---|
| Components |
|
-Supramolecule #1: Complex of Rvb1 and Rvb2 together with Tah1 and Pih1
| Supramolecule | Name: Complex of Rvb1 and Rvb2 together with Tah1 and Pih1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 358 KDa |
-Macromolecule #1: Rvb1p
| Macromolecule | Name: Rvb1p / type: other / ID: 1 / Classification: other |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MVAISEVKEN PGVNSSNSGA VTRTAAHTHI KGLGLDESGV AKRVEGGFVG QIEAREACGV IVDLIKAKKM SGRAILLAG GPSTGKTALA LAISQELGPK VPFCPLVGSE LYSVEVKKTE TLMENFRRAI GLRIKETKEV Y EGEVTELT PEDAENPLGG YGKTISHVIV ...String: MVAISEVKEN PGVNSSNSGA VTRTAAHTHI KGLGLDESGV AKRVEGGFVG QIEAREACGV IVDLIKAKKM SGRAILLAG GPSTGKTALA LAISQELGPK VPFCPLVGSE LYSVEVKKTE TLMENFRRAI GLRIKETKEV Y EGEVTELT PEDAENPLGG YGKTISHVIV GLKSAKGTKT LRLDPTIYES IQREKVSIGD VIYIEANTGA VK RVGRSDA YATEFDLETE EYVPLPKGEV HKKKEIVQDV TLHDLDVANA RPQGGQDVIS MMGQLLKPKK TEI TEKLRQ EVNKVVAKYI DQGVAELIPG VLFIDEVNML DIEIFTYLNK ALESNIAPVV VLASNRGMTT VRGT EDVIS PHGVPPDLID RLLIVRTLPY DKDEIRTIIE RRATVERLQV ESSALDLLAT MGTETSLRYA LQLLA PCGI LAQTSNRKEI VVNDVNEAKL LFLDAKRSTK ILETSANYL |
| Recombinant expression | Organism:  |
-Macromolecule #2: Rvb2p
| Macromolecule | Name: Rvb2p / type: other / ID: 2 / Classification: other |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MSIQTSDPNE TSDLKSLSLI AAHSHITGLG LDENLQPRPT SEGMVGQLQA RRAAGVILKM VQNGTIAGRA VLVAGPPST GKTALAMGVS QSLGKDVPFT AIAGSEIFSL ELSKTEALTQ AFRKSIGIKI KEETELIEGE V VEIQIDRS ITGGHKQGKL TIKTTDMETI ...String: MSIQTSDPNE TSDLKSLSLI AAHSHITGLG LDENLQPRPT SEGMVGQLQA RRAAGVILKM VQNGTIAGRA VLVAGPPST GKTALAMGVS QSLGKDVPFT AIAGSEIFSL ELSKTEALTQ AFRKSIGIKI KEETELIEGE V VEIQIDRS ITGGHKQGKL TIKTTDMETI YELGNKMIDG LTKEKVLAGD VISIDKASGK ITKLGRSFAR SR DYDAMGA DTRFVQCPEG ELQKRKTVVH TVSLHEIDVI NSRTQGFLAL FTGDTGEIRS EVRDQINTKV AEW KEEGKA EIVPGVLFID EVHMLDIECF SFINRALEDE FAPIVMMATN RGVSKTRGTN YKSPHGLPLD LLDR SIIIT TKSYNEQEIK TILSIRAQEE EVELSSDALD LLTKTGVETS LRYSSNLISV AQQIAMKRKN NTVEV EDVK RAYLLFLDSA RSVKYVQENE SQYIDDQGNV QISIAKSADP DAMDTTE |
| Recombinant expression | Organism:  |
-Macromolecule #3: Pih1p
| Macromolecule | Name: Pih1p / type: other / ID: 3 / Classification: other |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MADFLLRPIK QRHRNEDKYV SVDAADGSVS KIEPIADFVI KTKLLSANGP EKLQDGRKVF INVCHSPLVP KPEVDFNAR IVFPLIIQNE WEIPIITSCY RMDHDKKGQE CYVWDCCINS DCSRWICDDI QLREILVEWC L ESCEIRDS VVLCRDRIAF PKMKKKGAEL ...String: MADFLLRPIK QRHRNEDKYV SVDAADGSVS KIEPIADFVI KTKLLSANGP EKLQDGRKVF INVCHSPLVP KPEVDFNAR IVFPLIIQNE WEIPIITSCY RMDHDKKGQE CYVWDCCINS DCSRWICDDI QLREILVEWC L ESCEIRDS VVLCRDRIAF PKMKKKGAEL PALEVLNDEL HQDYKAKMHK IIEEEAGDPM SILRGRNDDG DD NNDPDDG TLPPLFPIEN KISGAKIEEI DKNEIAHRNL KQAPAPAPAP HEQQEDVPEY EVKMKRFKGA AYK LRILIE NKAPNSKPDR FSPSYNFAEN ILYINGKLSI PLPRDIVVNA ADIKIFHIRK ERTLYIYI |
| Recombinant expression | Organism:  |
-Macromolecule #4: Tah1p
| Macromolecule | Name: Tah1p / type: other / ID: 4 / Classification: other |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MSQFEKQKEQ GNSLFKQGLY REAVHCYDQL ITAQPQNPVG YSNKAMALIK LGEYTQAIQM CQQGLRYTST AEHVAIRSK LQYRLELAQG AVGSVQIPVV EVDELPEGYD RS |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 83.0 K / Max: 93.0 K |
| Image recording | Film or detector model: DIRECT ELECTRON DE-20 (5k x 3k) / Detector mode: INTEGRATING / Number real images: 1396 / Average electron dose: 58.68 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: -3.0 µm / Calibrated defocus min: -1.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: -3.0 µm / Nominal defocus min: -1.0 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)