+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8799 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Yeast 80S ribosome derived from EMPIAR-10045 using emClarity | |||||||||
 Map data Map data | Yeast 80S ribosome derived from EMPIAR-10045 using emClarity | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
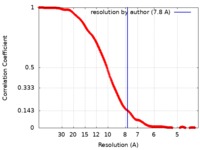
| Method | subtomogram averaging / cryo EM / Resolution: 7.8 Å | |||||||||
 Authors Authors | Himes BA / Zhang P | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Methods / Year: 2018 Journal: Nat Methods / Year: 2018Title: emClarity: software for high-resolution cryo-electron tomography and subtomogram averaging. Authors: Benjamin A Himes / Peijun Zhang /   Abstract: Macromolecular complexes are intrinsically flexible and often challenging to purify for structure determination by single-particle cryo-electron microscopy (cryo-EM). Such complexes can be studied by ...Macromolecular complexes are intrinsically flexible and often challenging to purify for structure determination by single-particle cryo-electron microscopy (cryo-EM). Such complexes can be studied by cryo-electron tomography (cryo-ET) combined with subtomogram alignment and classification, which in exceptional cases achieves subnanometer resolution, yielding insight into structure-function relationships. However, it remains challenging to apply this approach to specimens that exhibit conformational or compositional heterogeneity or are present in low abundance. To address this, we developed emClarity ( https://github.com/bHimes/emClarity/wiki ), a GPU-accelerated image-processing package featuring an iterative tomographic tilt-series refinement algorithm that uses subtomograms as fiducial markers and a 3D-sampling-function-compensated, multi-scale principal component analysis classification method. We demonstrate that our approach offers substantial improvement in the resolution of maps and in the separation of different functional states of macromolecular complexes compared with current state-of-the-art software. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8799.map.gz emd_8799.map.gz | 13.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8799-v30.xml emd-8799-v30.xml emd-8799.xml emd-8799.xml | 11.6 KB 11.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8799_fsc.xml emd_8799_fsc.xml | 34.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_8799.png emd_8799.png | 117.3 KB | ||
| Masks |  emd_8799_msk_1.map emd_8799_msk_1.map emd_8799_msk_2.map emd_8799_msk_2.map | 115.9 MB 115.9 MB |  Mask map Mask map | |
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8799 http://ftp.pdbj.org/pub/emdb/structures/EMD-8799 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8799 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8799 | HTTPS FTP |
-Validation report
| Summary document |  emd_8799_validation.pdf.gz emd_8799_validation.pdf.gz | 79.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_8799_full_validation.pdf.gz emd_8799_full_validation.pdf.gz | 78.3 KB | Display | |
| Data in XML |  emd_8799_validation.xml.gz emd_8799_validation.xml.gz | 495 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8799 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8799 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8799 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8799 | HTTPS FTP |
-Related structure data
| Related structure data |  8802C  8803C  8804C  8805C  8806C  8986C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8799.map.gz / Format: CCP4 / Size: 115.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8799.map.gz / Format: CCP4 / Size: 115.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Yeast 80S ribosome derived from EMPIAR-10045 using emClarity | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.17 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_8799_msk_1.map emd_8799_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Mask #2
| File |  emd_8799_msk_2.map emd_8799_msk_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : S. cerevisiae 80S ribosome
| Entire | Name: S. cerevisiae 80S ribosome |
|---|---|
| Components |
|
-Supramolecule #1: S. cerevisiae 80S ribosome
| Supramolecule | Name: S. cerevisiae 80S ribosome / type: complex / ID: 1 / Parent: 0 / Details: Non-translating yeast 80s ribosome |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER/RHODIUM / Mesh: 200 / Support film - Material: CARBON / Support film - topology: CONTINUOUS |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: Gatan Quantum Energy Filter |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Average electron dose: 2.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 3.0 µm / Nominal magnification: 53000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)