+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8796 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Katanin hexamer in the ring conformation | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | microtubule cytoskeleton / microtubule severing protein / AAA ATPase / p60 / MOTOR PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of meiotic spindle elongation / MATH domain binding / striated muscle myosin thick filament assembly / meiotic spindle disassembly / katanin complex / meiotic spindle pole / microtubule-severing ATPase / microtubule severing ATPase activity / microtubule severing / female meiotic nuclear division ...negative regulation of meiotic spindle elongation / MATH domain binding / striated muscle myosin thick filament assembly / meiotic spindle disassembly / katanin complex / meiotic spindle pole / microtubule-severing ATPase / microtubule severing ATPase activity / microtubule severing / female meiotic nuclear division / meiotic spindle organization / embryo development ending in birth or egg hatching / meiotic spindle / microtubule depolymerization / spindle / spindle pole / protein phosphatase binding / microtubule binding / microtubule / molecular adaptor activity / cell division / centrosome / chromatin / ATP hydrolysis activity / ATP binding / identical protein binding / nucleus / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
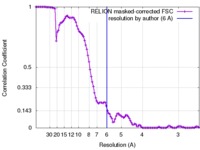
| Method | single particle reconstruction / cryo EM / Resolution: 6.0 Å | |||||||||
 Authors Authors | Zehr EA / Szyk A | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2017 Journal: Nat Struct Mol Biol / Year: 2017Title: Katanin spiral and ring structures shed light on power stroke for microtubule severing. Authors: Elena Zehr / Agnieszka Szyk / Grzegorz Piszczek / Ewa Szczesna / Xiaobing Zuo / Antonina Roll-Mecak /  Abstract: Microtubule-severing enzymes katanin, spastin and fidgetin are AAA ATPases important for the biogenesis and maintenance of complex microtubule arrays in axons, spindles and cilia. Because of a lack ...Microtubule-severing enzymes katanin, spastin and fidgetin are AAA ATPases important for the biogenesis and maintenance of complex microtubule arrays in axons, spindles and cilia. Because of a lack of known 3D structures for these enzymes, their mechanism of action has remained poorly understood. Here we report the X-ray crystal structure of the monomeric AAA katanin module from Caenorhabditis elegans and cryo-EM reconstructions of the hexamer in two conformations. The structures reveal an unexpected asymmetric arrangement of the AAA domains mediated by structural elements unique to microtubule-severing enzymes and critical for their function. The reconstructions show that katanin cycles between open spiral and closed ring conformations, depending on the ATP occupancy of a gating protomer that tenses or relaxes interprotomer interfaces. Cycling of the hexamer between these conformations would provide the power stroke for microtubule severing. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8796.map.gz emd_8796.map.gz | 5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8796-v30.xml emd-8796-v30.xml emd-8796.xml emd-8796.xml | 18.2 KB 18.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8796_fsc.xml emd_8796_fsc.xml | 10.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_8796.png emd_8796.png | 111.3 KB | ||
| Filedesc metadata |  emd-8796.cif.gz emd-8796.cif.gz | 6.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8796 http://ftp.pdbj.org/pub/emdb/structures/EMD-8796 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8796 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8796 | HTTPS FTP |
-Related structure data
| Related structure data |  5wcbMC  8794C  5wc0C  5wc1C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8796.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8796.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.31 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : katanin
| Entire | Name: katanin |
|---|---|
| Components |
|
-Supramolecule #1: katanin
| Supramolecule | Name: katanin / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 310 KDa |
-Macromolecule #1: Meiotic spindle formation protein mei-1
| Macromolecule | Name: Meiotic spindle formation protein mei-1 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number: ec: 3.6.4.3 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 51.802359 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNGDVQSVIR GYLERAQVAK TMSDAGRWNE AGDLLRQLMT DVKSCKISAS NRDEHDARNT FLRALEANLK LVQQNVRDED DLHEAMTRQ SGSPEPPADP DVWSKPSPPL PSSSKFGATK KGVGAAGPRP REISKSTSSM STNPADVKPA NPTQGILPQN S AGDSFDAS ...String: MNGDVQSVIR GYLERAQVAK TMSDAGRWNE AGDLLRQLMT DVKSCKISAS NRDEHDARNT FLRALEANLK LVQQNVRDED DLHEAMTRQ SGSPEPPADP DVWSKPSPPL PSSSKFGATK KGVGAAGPRP REISKSTSSM STNPADVKPA NPTQGILPQN S AGDSFDAS AYDAYIVQAV RGTMATNTEN TMSLDDIIGM HDVKQVLHEA VTLPLLVPEF FQGLRSPWKA MVLAGPPGTG KT LIARAIA SESSSTFFTV SSTDLSSKWR GDSEKIVRLL FELARFYAPS IIFIDQIDTL GGQRGNSGEH EASRRVKSEF LVQ MDGSQN KFDSRRVFVL AATNIPWELD EALRRRFEKR IFIPLPDIDA RKKLIEKSME GTPKSDEINY DDLAARTEGF SGAD VVSLC RTAAINVLRR YDTKSLRGGE LTAAMESLKA ELVRNIDFEA ALQAVSPSAG PDTMLKCKEW CDSFGAM UniProtKB: Meiotic spindle formation protein mei-1 |
-Macromolecule #2: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 5 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.6 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: C-flat / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 2 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 6 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 298 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3837 pixel / Digitization - Dimensions - Height: 3710 pixel / Number grids imaged: 1 / Number real images: 2100 / Average exposure time: 17.5 sec. / Average electron dose: 51.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 22500 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)