[English] 日本語
 Yorodumi
Yorodumi- EMDB-8702: Structure of a mechanotransduction ion channel Drosophila NOMPC i... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8702 | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of a mechanotransduction ion channel Drosophila NOMPC in nanodisc | ||||||||||||||||||||||||
 Map data Map data | an ion channel in nano disc -- final sharpened map | ||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords | Membrane protein / Mechanotransduction Ion channel | ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationsensory perception of touch / detection of mechanical stimulus involved in sensory perception of touch / detection of mechanical stimulus involved in sensory perception / mechanosensory behavior / sensory perception of mechanical stimulus / response to auditory stimulus / mechanosensitive monoatomic ion channel activity / cation channel complex / locomotion / ankyrin binding ...sensory perception of touch / detection of mechanical stimulus involved in sensory perception of touch / detection of mechanical stimulus involved in sensory perception / mechanosensory behavior / sensory perception of mechanical stimulus / response to auditory stimulus / mechanosensitive monoatomic ion channel activity / cation channel complex / locomotion / ankyrin binding / startle response / monoatomic ion channel activity / monoatomic cation transport / monoatomic cation channel activity / sensory perception of sound / cellular response to mechanical stimulus / calcium channel activity / calcium ion transport / cilium / neuronal cell body / dendrite / plasma membrane Similarity search - Function | ||||||||||||||||||||||||
| Biological species |  | ||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.55 Å | ||||||||||||||||||||||||
 Authors Authors | Jin P | ||||||||||||||||||||||||
| Funding support |  United States, 7 items United States, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2017 Journal: Nature / Year: 2017Title: Electron cryo-microscopy structure of the mechanotransduction channel NOMPC. Authors: Peng Jin / David Bulkley / Yanmeng Guo / Wei Zhang / Zhenhao Guo / Walter Huynh / Shenping Wu / Shan Meltzer / Tong Cheng / Lily Yeh Jan / Yuh-Nung Jan / Yifan Cheng /  Abstract: Mechanosensory transduction for senses such as proprioception, touch, balance, acceleration, hearing and pain relies on mechanotransduction channels, which convert mechanical stimuli into electrical ...Mechanosensory transduction for senses such as proprioception, touch, balance, acceleration, hearing and pain relies on mechanotransduction channels, which convert mechanical stimuli into electrical signals in specialized sensory cells. How force gates mechanotransduction channels is a central question in the field, for which there are two major models. One is the membrane-tension model: force applied to the membrane generates a change in membrane tension that is sufficient to gate the channel, as in the bacterial MscL channel and certain eukaryotic potassium channels. The other is the tether model: force is transmitted via a tether to gate the channel. The transient receptor potential (TRP) channel NOMPC is important for mechanosensation-related behaviours such as locomotion, touch and sound sensation across different species including Caenorhabditis elegans, Drosophila and zebrafish. NOMPC is the founding member of the TRPN subfamily, and is thought to be gated by tethering of its ankyrin repeat domain to microtubules of the cytoskeleton. Thus, a goal of studying NOMPC is to reveal the underlying mechanism of force-induced gating, which could serve as a paradigm of the tether model. NOMPC fulfils all the criteria that apply to mechanotransduction channels and has 29 ankyrin repeats, the largest number among TRP channels. A key question is how the long ankyrin repeat domain is organized as a tether that can trigger channel gating. Here we present a de novo atomic structure of Drosophila NOMPC determined by single-particle electron cryo-microscopy. Structural analysis suggests that the ankyrin repeat domain of NOMPC resembles a helical spring, suggesting its role of linking mechanical displacement of the cytoskeleton to the opening of the channel. The NOMPC architecture underscores the basis of translating mechanical force into an electrical signal within a cell. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8702.map.gz emd_8702.map.gz | 6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8702-v30.xml emd-8702-v30.xml emd-8702.xml emd-8702.xml | 31.1 KB 31.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8702.png emd_8702.png | 49.3 KB | ||
| Filedesc metadata |  emd-8702.cif.gz emd-8702.cif.gz | 7.4 KB | ||
| Others |  emd_8702_additional_1.map.gz emd_8702_additional_1.map.gz emd_8702_additional_2.map.gz emd_8702_additional_2.map.gz emd_8702_additional_3.map.gz emd_8702_additional_3.map.gz emd_8702_additional_4.map.gz emd_8702_additional_4.map.gz emd_8702_half_map_1.map.gz emd_8702_half_map_1.map.gz emd_8702_half_map_2.map.gz emd_8702_half_map_2.map.gz | 215.6 MB 4.8 MB 5 MB 6.7 MB 211.5 MB 211.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8702 http://ftp.pdbj.org/pub/emdb/structures/EMD-8702 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8702 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8702 | HTTPS FTP |
-Related structure data
| Related structure data |  5vkqMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10093 (Title: Structure of an ion channel in nano disc / Data size: 203.9 EMPIAR-10093 (Title: Structure of an ion channel in nano disc / Data size: 203.9 Data #1: Motion corrected dose weighted summed frames [micrographs - single frame] Data #2: Motion corrected dose weighted particle images [picked particles - multiframe - processed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8702.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8702.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | an ion channel in nano disc -- final sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2156 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: an ion channel in nano disc -- final unsharpened map
| File | emd_8702_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | an ion channel in nano disc -- final unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
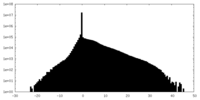
| Density Histograms |
-Additional map: an ion channel in nano disc -- class 1
| File | emd_8702_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | an ion channel in nano disc -- class 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: an ion channel in nano disc -- class 2
| File | emd_8702_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | an ion channel in nano disc -- class 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
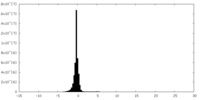
| Density Histograms |
-Additional map: an ion channel in nano disc -- class 3
| File | emd_8702_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | an ion channel in nano disc -- class 3 | ||||||||||||
| Projections & Slices |
| ||||||||||||
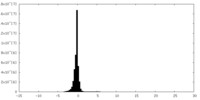
| Density Histograms |
-Half map: an ion channel in nano disc -- half volume 1
| File | emd_8702_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | an ion channel in nano disc -- half volume 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
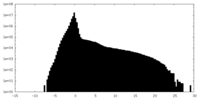
| Density Histograms |
-Half map: an ion channel in nano disc -- half volume 2
| File | emd_8702_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | an ion channel in nano disc -- half volume 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Tetrameric channel
| Entire | Name: Tetrameric channel |
|---|---|
| Components |
|
-Supramolecule #1: Tetrameric channel
| Supramolecule | Name: Tetrameric channel / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 754.893 KDa |
-Macromolecule #1: No mechanoreceptor potential C isoform L
| Macromolecule | Name: No mechanoreceptor potential C isoform L / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 188.978734 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSQPRGGRGG GRGGGVGRKT PSSLTGPPDE SATPSERATP ASKADSDPKD DSSSNGDKKD MDLFPAPKPP SAGASIRDTA NKVLGLAMK SEWTPIEAEL KKLEKYVANV GEDGNHIPLA GVHDMNTGMT PLMYATKDNK TAIMDRMIEL GADVGARNND N YNVLHIAA ...String: MSQPRGGRGG GRGGGVGRKT PSSLTGPPDE SATPSERATP ASKADSDPKD DSSSNGDKKD MDLFPAPKPP SAGASIRDTA NKVLGLAMK SEWTPIEAEL KKLEKYVANV GEDGNHIPLA GVHDMNTGMT PLMYATKDNK TAIMDRMIEL GADVGARNND N YNVLHIAA MYSREDVVKL LLTKRGVDPF STGGSRSQTA VHLVSSRQTG TATNILRALL AAAGKDIRLK ADGRGKIPLL LA VESGNQS MCRELLAAQT AEQLKATTAN GDTALHLAAR RRDVDMVRIL VDYGTNVDTQ NGEGQTPLHI AAAEGDEALL KYF YGVRAS ASIADNQDRT PMHLAAENGH AHVIEILADK FKASIFERTK DGSTLMHIAS LNGHAECATM LFKKGVYLHM PNKD GARSI HTAAAYGHTG IINTLLQKGE KVDVTTNDNY TALHIAVESA KPAVVETLLG FGADVHVRGG KLRETPLHIA ARVKD GDRC ALMLLKSGAS PNLTTDDCLT PVHVAARHGN LATLMQLLED EGDPLYKSNT GETPLHMACR ACHPDIVRHL IETVKE KHG PDKATTYINS VNEDGATALH YTCQITKEEV KIPESDKQIV RMLLENGADV TLQTKTALET AFHYCAVAGN NDVLMEM IS HMNPTDIQKA MNRQSSVGWT PLLIACHRGH MELVNNLLAN HARVDVFDTE GRSALHLAAE RGYLHVCDAL LTNKAFIN S KSRVGRTALH LAAMNGFTHL VKFLIKDHNA VIDILTLRKQ TPLHLAAASG QMEVCQLLLE LGANIDATDD LGQKPIHVA AQNNYSEVAK LFLQQHPSLV NATSKDGNTC AHIAAMQGSV KVIEELMKFD RSGVISARNK LTDATPLQLA AEGGHADVVK ALVRAGASC TEENKAGFTA VHLAAQNGHG QVLDVLKSTN SLRINSKKLG LTPLHVAAYY GQADTVRELL TSVPATVKSE T PTGQSLFG DLGTESGMTP LHLAAFSGNE NVVRLLLNSA GVQVDAATIE NGYNPLHLAC FGGHMSVVGL LLSRSAELLQ SQ DRNGRTG LHIAAMHGHI QMVEILLGQG AEINATDRNG WTPLHCAAKA GHLEVVKLLC EAGASPKSET NYGCAAIWFA ASE GHNEVL RYLMNKEHDT YGLMEDKRFV YNLMVVSKNH NNKPIQEFVL VSPAPVDTAA KLSNIYIVLS TKEKERAKDL VAAG KQCEA MATELLALAA GSDSAGKILQ ATDKRNVEFL DVLIENEQKE VIAHTVVQRY LQELWHGSLT WASWKILLLL VAFIV CPPV WIGFTFPMGH KFNKVPIIKF MSYLTSHIYL MIHLSIVGIT PIYPVLRLSL VPYWYEVGLL IWLSGLLLFE LTNPSD KSG LGSIKVLVLL LGMAGVGVHV SAFLFVSKEY WPTLVYCRNQ CFALAFLLAC VQILDFLSFH HLFGPWAIII GDLLKDL AR FLAVLAIFVF GFSMHIVALN QSFANFSPED LRSFEKKNRN RGYFSDVRMH PINSFELLFF AVFGQTTTEQ TQVDKIKN V ATPTQPYWVE YLFKIVFGIY MLVSVVVLIQ LLIAMMSDTY QRIQAQSDIE WKFGLSKLIR NMHRTTTAPS PLNLVTTWF MWIVEKVKAR MKKKKRPSLV QMMGIRQASP RTKAGAKWLS KIKKDSVALS QVHLSPLGSQ ASFSQANQNR IENVADWEAI AKKYRALVG DEEGGSLKDS DAESGSQEGS GGQQPPAQVG RRAIKATLAD TTK UniProtKB: No mechanoreceptor potential C isoform L |
-Macromolecule #2: 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE
| Macromolecule | Name: 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE / type: ligand / ID: 2 / Number of copies: 32 / Formula: PCF |
|---|---|
| Molecular weight | Theoretical: 734.039 Da |
| Chemical component information |  ChemComp-PCF: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.4 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8.5 Component:
| ||||||||||||
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 92 % / Chamber temperature: 20 K / Instrument: GATAN CRYOPLUNGE 3 Details: 2.5 sec blot, -1 offset, whatman #1 filter paper on front side, plastic on back side. | ||||||||||||
| Details | sample solubilized in nanodisc |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Temperature | Min: 83.0 K / Max: 83.0 K |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 2-40 / Number grids imaged: 1 / Number real images: 1873 / Average exposure time: 8.0 sec. / Average electron dose: 54.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 30.0 µm / Calibrated defocus max: -3.3000000000000003 µm / Calibrated defocus min: -1.4000000000000001 µm / Calibrated magnification: 41132 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.1 mm / Nominal defocus max: -3.0 µm / Nominal defocus min: -1.5 µm / Nominal magnification: 31000 |
| Sample stage | Specimen holder model: GATAN HCHST 3008 SINGLE TILT HIGH RESOLUTION HELIUM COOLING HOLDER Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 125.7 |
|---|---|
| Output model |  PDB-5vkq: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)