[English] 日本語
 Yorodumi
Yorodumi- EMDB-8637: CryoEM structure of the ERAD-associated E3 ubiquitin-protein liga... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8637 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of the ERAD-associated E3 ubiquitin-protein ligase HRD1 | |||||||||
 Map data Map data | Hrd1 dimer map filtered to 4.1A and sharpened with -230 b-factor | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Retrotranslocon / E3 ligase / ERAD / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationHrd1p ubiquitin ligase ERAD-M complex / Hrd1p ubiquitin ligase complex / Hrd1p ubiquitin ligase ERAD-L complex / fungal-type cell wall organization / retrograde protein transport, ER to cytosol / endoplasmic reticulum unfolded protein response / protein autoubiquitination / protein K48-linked ubiquitination / ERAD pathway / RING-type E3 ubiquitin transferase ...Hrd1p ubiquitin ligase ERAD-M complex / Hrd1p ubiquitin ligase complex / Hrd1p ubiquitin ligase ERAD-L complex / fungal-type cell wall organization / retrograde protein transport, ER to cytosol / endoplasmic reticulum unfolded protein response / protein autoubiquitination / protein K48-linked ubiquitination / ERAD pathway / RING-type E3 ubiquitin transferase / ubiquitin-protein transferase activity / ubiquitin protein ligase activity / ubiquitin-dependent protein catabolic process / endoplasmic reticulum membrane / endoplasmic reticulum / zinc ion binding / identical protein binding Similarity search - Function | |||||||||
| Biological species |   | |||||||||
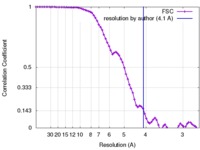
| Method | single particle reconstruction / cryo EM / Resolution: 4.1 Å | |||||||||
 Authors Authors | Schoebel S / Mi W | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2017 Journal: Nature / Year: 2017Title: Cryo-EM structure of the protein-conducting ERAD channel Hrd1 in complex with Hrd3. Authors: Stefan Schoebel / Wei Mi / Alexander Stein / Sergey Ovchinnikov / Ryan Pavlovicz / Frank DiMaio / David Baker / Melissa G Chambers / Huayou Su / Dongsheng Li / Tom A Rapoport / Maofu Liao /    Abstract: Misfolded endoplasmic reticulum proteins are retro-translocated through the membrane into the cytosol, where they are poly-ubiquitinated, extracted from the membrane, and degraded by the proteasome-a ...Misfolded endoplasmic reticulum proteins are retro-translocated through the membrane into the cytosol, where they are poly-ubiquitinated, extracted from the membrane, and degraded by the proteasome-a pathway termed endoplasmic reticulum-associated protein degradation (ERAD). Proteins with misfolded domains in the endoplasmic reticulum lumen or membrane are discarded through the ERAD-L and ERAD-M pathways, respectively. In Saccharomyces cerevisiae, both pathways require the ubiquitin ligase Hrd1, a multi-spanning membrane protein with a cytosolic RING finger domain. Hrd1 is the crucial membrane component for retro-translocation, but it is unclear whether it forms a protein-conducting channel. Here we present a cryo-electron microscopy structure of S. cerevisiae Hrd1 in complex with its endoplasmic reticulum luminal binding partner, Hrd3. Hrd1 forms a dimer within the membrane with one or two Hrd3 molecules associated at its luminal side. Each Hrd1 molecule has eight transmembrane segments, five of which form an aqueous cavity extending from the cytosol almost to the endoplasmic reticulum lumen, while a segment of the neighbouring Hrd1 molecule forms a lateral seal. The aqueous cavity and lateral gate are reminiscent of features of protein-conducting conduits that facilitate polypeptide movement in the opposite direction-from the cytosol into or across membranes. Our results suggest that Hrd1 forms a retro-translocation channel for the movement of misfolded polypeptides through the endoplasmic reticulum membrane. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8637.map.gz emd_8637.map.gz | 25 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8637-v30.xml emd-8637-v30.xml emd-8637.xml emd-8637.xml | 14 KB 14 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8637_fsc.xml emd_8637_fsc.xml | 8 KB | Display |  FSC data file FSC data file |
| Images |  emd_8637.png emd_8637.png | 166.6 KB | ||
| Filedesc metadata |  emd-8637.cif.gz emd-8637.cif.gz | 5.6 KB | ||
| Others |  emd_8637_additional.map.gz emd_8637_additional.map.gz | 20.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8637 http://ftp.pdbj.org/pub/emdb/structures/EMD-8637 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8637 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8637 | HTTPS FTP |
-Related structure data
| Related structure data |  5v6pMC  8638C  8639C  8642C  5v7vC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8637.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8637.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Hrd1 dimer map filtered to 4.1A and sharpened with -230 b-factor | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.35 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Hrd1 dimer map without filtering or amplitude modification
| File | emd_8637_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Hrd1 dimer map without filtering or amplitude modification | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Hrd1 dimer in Hrd1/Hrd3 complex
| Entire | Name: Hrd1 dimer in Hrd1/Hrd3 complex |
|---|---|
| Components |
|
-Supramolecule #1: Hrd1 dimer in Hrd1/Hrd3 complex
| Supramolecule | Name: Hrd1 dimer in Hrd1/Hrd3 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: ERAD-associated E3 ubiquitin-protein ligase HRD1
| Macromolecule | Name: ERAD-associated E3 ubiquitin-protein ligase HRD1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: RING-type E3 ubiquitin transferase |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 47.866418 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MVPENRRKQL AIFVVVTYLL TFYCVYSATK TSVSFLQVTL KLNEGFNLMV LSIFILLNST LLWQLLTKLL FGELRLIEHE HIFERLPFT IINTLFMSSL FHERYFFTVA FFGLLLLYLK VFHWILKDRL EALLQSINDS TTMKTLIFSR FSFNLVLLAV V DYQIITRC ...String: MVPENRRKQL AIFVVVTYLL TFYCVYSATK TSVSFLQVTL KLNEGFNLMV LSIFILLNST LLWQLLTKLL FGELRLIEHE HIFERLPFT IINTLFMSSL FHERYFFTVA FFGLLLLYLK VFHWILKDRL EALLQSINDS TTMKTLIFSR FSFNLVLLAV V DYQIITRC ISSIYTNQKS DIESTSLYLI QVMEFTMLLI DLLNLFLQTC LNFWEFYRSQ QSLSNENNHI VHGDPTDENT VE SDQSQPV LNDDDDDDDD DRQFTGLEGK FMYEKAIDVF TRFLKTALHL SMLIPFRMPM MLLKDVVWDI LALYQSGTSL WKI WRNNKQ LDDTLVTVTV EQLQNSANDD NICIICMDEL IHSPNQQTWK NKNKKPKRLP CGHILHLSCL KNWMERSQTC PICR LPVFD EK UniProtKB: ERAD-associated E3 ubiquitin-protein ligase HRD1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.8 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 82.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)