[English] 日本語
 Yorodumi
Yorodumi- EMDB-8441: Structure of bL17-depleted large ribosomal subunit assembly inter... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8441 | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of bL17-depleted large ribosomal subunit assembly intermediate - Class C | ||||||||||||||||||||||||
 Map data Map data | Class C bL17-Depleted 50S Ribosomal Biogenesis Intermediate | ||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
| Biological species |  | ||||||||||||||||||||||||
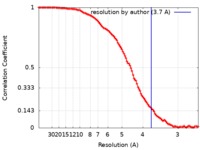
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | ||||||||||||||||||||||||
 Authors Authors | Tan Y / Davis JH / Lyumkis D | ||||||||||||||||||||||||
| Funding support |  United States, United States,  Singapore, 7 items Singapore, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Cell / Year: 2016 Journal: Cell / Year: 2016Title: Modular Assembly of the Bacterial Large Ribosomal Subunit. Authors: Joseph H Davis / Yong Zi Tan / Bridget Carragher / Clinton S Potter / Dmitry Lyumkis / James R Williamson /  Abstract: The ribosome is a complex macromolecular machine and serves as an ideal system for understanding biological macromolecular assembly. Direct observation of ribosome assembly in vivo is difficult, as ...The ribosome is a complex macromolecular machine and serves as an ideal system for understanding biological macromolecular assembly. Direct observation of ribosome assembly in vivo is difficult, as few intermediates have been isolated and thoroughly characterized. Herein, we deploy a genetic system to starve cells of an essential ribosomal protein, which results in the accumulation of assembly intermediates that are competent for maturation. Quantitative mass spectrometry and single-particle cryo-electron microscopy reveal 13 distinct intermediates, which were each resolved to ∼4-5 Å resolution and could be placed in an assembly pathway. We find that ribosome biogenesis is a parallel process, that blocks of structured rRNA and proteins assemble cooperatively, and that the entire process is dynamic and can be "re-routed" through different pathways as needed. This work reveals the complex landscape of ribosome assembly in vivo and provides the requisite tools to characterize additional assembly pathways for ribosomes and other macromolecular machines. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8441.map.gz emd_8441.map.gz | 102.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8441-v30.xml emd-8441-v30.xml emd-8441.xml emd-8441.xml | 25.8 KB 25.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8441_fsc.xml emd_8441_fsc.xml | 11.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_8441.png emd_8441.png | 119.8 KB | ||
| Masks |  emd_8441_msk_1.map emd_8441_msk_1.map | 125 MB |  Mask map Mask map | |
| Others |  emd_8441_additional.map.gz emd_8441_additional.map.gz emd_8441_half_map_1.map.gz emd_8441_half_map_1.map.gz emd_8441_half_map_2.map.gz emd_8441_half_map_2.map.gz | 20.2 MB 20.1 MB 20.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8441 http://ftp.pdbj.org/pub/emdb/structures/EMD-8441 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8441 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8441 | HTTPS FTP |
-Related structure data
| Related structure data |  8434C  8440C  8442C  8443C  8444C  8445C  8446C  8447C  8448C  8449C  8450C  8451C  8452C  8453C  8455C  8456C  8457C C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10076 (Title: CryoEM Dataset of L17-Depleted 50S Ribosomal Intermediates EMPIAR-10076 (Title: CryoEM Dataset of L17-Depleted 50S Ribosomal IntermediatesData size: 50.3 Data #1: Particle stack of L17-depleted 50S Ribosomal Assembly Intermediates [picked particles - multiframe - unprocessed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8441.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8441.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Class C bL17-Depleted 50S Ribosomal Biogenesis Intermediate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.31 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
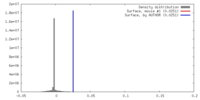
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
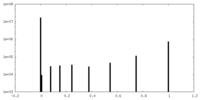
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_8441_msk_1.map emd_8441_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
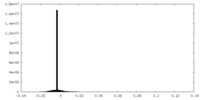
| Density Histograms |
-Additional map: Class C bL17-Depleted 50S Ribosomal Biogenesis Intermediate -...
| File | emd_8441_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Class C bL17-Depleted 50S Ribosomal Biogenesis Intermediate - Unsharpened | ||||||||||||
| Projections & Slices |
| ||||||||||||
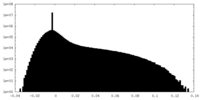
| Density Histograms |
-Half map: Class C bL17-Depleted 50S Ribosomal Biogenesis Intermediate -...
| File | emd_8441_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Class C bL17-Depleted 50S Ribosomal Biogenesis Intermediate - Halfmap 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Class C bL17-Depleted 50S Ribosomal Biogenesis Intermediate -...
| File | emd_8441_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Class C bL17-Depleted 50S Ribosomal Biogenesis Intermediate - Halfmap 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : bL17-depleted large ribosomal subunit assembly intermediate - Class C
| Entire | Name: bL17-depleted large ribosomal subunit assembly intermediate - Class C |
|---|---|
| Components |
|
-Supramolecule #1: bL17-depleted large ribosomal subunit assembly intermediate - Class C
| Supramolecule | Name: bL17-depleted large ribosomal subunit assembly intermediate - Class C type: complex / ID: 1 / Parent: 0 Details: E. coli large ribosome subunit purified from bL17-depleted cells. |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
Details: Most of the sucrose was removed by spin concentration | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: C-Flat CF-1.2/1.3-4Au / Material: GOLD / Mesh: 400 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Atmosphere: OTHER | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 298 K / Instrument: GATAN CRYOPLUNGE 3 / Details: 3 microliter of the sample was added.. | ||||||||||||||||||
| Details | Fraction was obtained directly from a sucrose gradient, and was spin-concentrated in a 100 kDa MW-cutoff filter. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Details | In order to account for highly preferred orientation of the specimen, data was acquired using tilts ranging from 10-60 degrees, in addition to untilted images at 0 degrees. The CTF was estimated on a per-particle basis to account for the gradient of CTF values across individual micrographs |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Digitization - Sampling interval: 5.0 µm / Digitization - Frames/image: 1-50 / Number grids imaged: 2 / Number real images: 2479 / Average exposure time: 10.0 sec. / Average electron dose: 34.27 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated magnification: 38167 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 22500 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)