[English] 日本語
 Yorodumi
Yorodumi- EMDB-8404: Single particle helical reconstruction of protease cleavage produ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8404 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Single particle helical reconstruction of protease cleavage product from HIV-1 Gag VLPs | ||||||||||||
 Map data Map data | CA tube assembled from gag cleavage | ||||||||||||
 Sample Sample |
| ||||||||||||
| Method | helical reconstruction / cryo EM / Resolution: 22.0 Å | ||||||||||||
 Authors Authors | Fu X / Zhang P | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2016 Journal: Nat Commun / Year: 2016Title: In vitro protease cleavage and computer simulations reveal the HIV-1 capsid maturation pathway. Authors: Jiying Ning / Gonca Erdemci-Tandogan / Ernest L Yufenyuy / Jef Wagner / Benjamin A Himes / Gongpu Zhao / Christopher Aiken / Roya Zandi / Peijun Zhang /   Abstract: HIV-1 virions assemble as immature particles containing Gag polyproteins that are processed by the viral protease into individual components, resulting in the formation of mature infectious particles. ...HIV-1 virions assemble as immature particles containing Gag polyproteins that are processed by the viral protease into individual components, resulting in the formation of mature infectious particles. There are two competing models for the process of forming the mature HIV-1 core: the disassembly and de novo reassembly model and the non-diffusional displacive model. To study the maturation pathway, we simulate HIV-1 maturation in vitro by digesting immature particles and assembled virus-like particles with recombinant HIV-1 protease and monitor the process with biochemical assays and cryoEM structural analysis in parallel. Processing of Gag in vitro is accurate and efficient and results in both soluble capsid protein and conical or tubular capsid assemblies, seemingly converted from immature Gag particles. Computer simulations further reveal probable assembly pathways of HIV-1 capsid formation. Combining the experimental data and computer simulations, our results suggest a sequential combination of both displacive and disassembly/reassembly processes for HIV-1 maturation. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8404.map.gz emd_8404.map.gz | 2.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8404-v30.xml emd-8404-v30.xml emd-8404.xml emd-8404.xml | 12.7 KB 12.7 KB | Display Display |  EMDB header EMDB header |
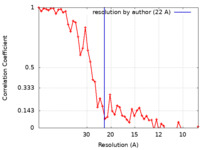
| FSC (resolution estimation) |  emd_8404_fsc.xml emd_8404_fsc.xml | 5.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_8404.png emd_8404.png | 203.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8404 http://ftp.pdbj.org/pub/emdb/structures/EMD-8404 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8404 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8404 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8404.map.gz / Format: CCP4 / Size: 8.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8404.map.gz / Format: CCP4 / Size: 8.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CA tube assembled from gag cleavage | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.52 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Single particle helical reconstruction of protease cleavage produ...
| Entire | Name: Single particle helical reconstruction of protease cleavage product from HIV-1 Gag VLPs |
|---|---|
| Components |
|
-Supramolecule #1: Single particle helical reconstruction of protease cleavage produ...
| Supramolecule | Name: Single particle helical reconstruction of protease cleavage product from HIV-1 Gag VLPs type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Concentration | 2.0 mg/mL |
|---|---|
| Buffer | pH: 6 / Component - Concentration: 50.0 mM / Component - Formula: NaAc / Component - Name: Sodium Acetate |
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: HOMEMADE PLUNGER |
| Details | sample is resulted from HIV-1 protease cleavage of in vitro assembled Gag VLPs |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Specialist optics | Spherical aberration corrector: No / Chromatic aberration corrector: No / Energy filter - Name: No |
| Image recording | Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Digitization - Sampling interval: 15.0 µm / Number grids imaged: 1 / Number real images: 2 / Average exposure time: 1.0 sec. / Average electron dose: 10.0 e/Å2 Details: Image was collected by single exposure, not movie mode. |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 9.0 µm / Calibrated defocus min: 8.0 µm / Calibrated magnification: 66262 / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal magnification: 50000 |
| Sample stage | Specimen holder model: GATAN 626 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)