[English] 日本語
 Yorodumi
Yorodumi- EMDB-8315: Structure of the STRA6 receptor for retinol uptake in complex wit... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8315 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the STRA6 receptor for retinol uptake in complex with calmodulin | |||||||||
 Map data Map data | Reconstruction of STRA6/CaM complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Vitamin A / retinol / STRA6 / membrane / MEMBRANE PROTEIN-CALCIUM BINDING PROTEIN complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationvitamin A import into cell / retinol transport / retinol transmembrane transporter activity / chordate embryonic development / retinal binding / retinol binding / enzyme regulator activity / signaling receptor activity / molecular adaptor activity / calmodulin binding ...vitamin A import into cell / retinol transport / retinol transmembrane transporter activity / chordate embryonic development / retinal binding / retinol binding / enzyme regulator activity / signaling receptor activity / molecular adaptor activity / calmodulin binding / calcium ion binding / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Clarke OB / Chen Y | |||||||||
 Citation Citation |  Journal: Science / Year: 2016 Journal: Science / Year: 2016Title: Structure of the STRA6 receptor for retinol uptake. Authors: Yunting Chen / Oliver B Clarke / Jonathan Kim / Sean Stowe / Youn-Kyung Kim / Zahra Assur / Michael Cavalier / Raquel Godoy-Ruiz / Desiree C von Alpen / Chiara Manzini / William S Blaner / ...Authors: Yunting Chen / Oliver B Clarke / Jonathan Kim / Sean Stowe / Youn-Kyung Kim / Zahra Assur / Michael Cavalier / Raquel Godoy-Ruiz / Desiree C von Alpen / Chiara Manzini / William S Blaner / Joachim Frank / Loredana Quadro / David J Weber / Lawrence Shapiro / Wayne A Hendrickson / Filippo Mancia /  Abstract: Vitamin A homeostasis is critical to normal cellular function. Retinol-binding protein (RBP) is the sole specific carrier in the bloodstream for hydrophobic retinol, the main form in which vitamin A ...Vitamin A homeostasis is critical to normal cellular function. Retinol-binding protein (RBP) is the sole specific carrier in the bloodstream for hydrophobic retinol, the main form in which vitamin A is transported. The integral membrane receptor STRA6 mediates cellular uptake of vitamin A by recognizing RBP-retinol to trigger release and internalization of retinol. We present the structure of zebrafish STRA6 determined to 3.9-angstrom resolution by single-particle cryo-electron microscopy. STRA6 has one intramembrane and nine transmembrane helices in an intricate dimeric assembly. Unexpectedly, calmodulin is bound tightly to STRA6 in a noncanonical arrangement. Residues involved with RBP binding map to an archlike structure that covers a deep lipophilic cleft. This cleft is open to the membrane, suggesting a possible mode for internalization of retinol through direct diffusion into the lipid bilayer. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8315.map.gz emd_8315.map.gz | 28.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8315-v30.xml emd-8315-v30.xml emd-8315.xml emd-8315.xml | 17.6 KB 17.6 KB | Display Display |  EMDB header EMDB header |
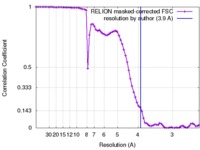
| FSC (resolution estimation) |  emd_8315_fsc.xml emd_8315_fsc.xml | 7.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_8315.png emd_8315.png | 147.5 KB | ||
| Masks |  emd_8315_msk_1.map emd_8315_msk_1.map | 30.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-8315.cif.gz emd-8315.cif.gz | 6.3 KB | ||
| Others |  emd_8315_half_map_1.map.gz emd_8315_half_map_1.map.gz emd_8315_half_map_2.map.gz emd_8315_half_map_2.map.gz | 22.5 MB 22.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8315 http://ftp.pdbj.org/pub/emdb/structures/EMD-8315 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8315 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8315 | HTTPS FTP |
-Related structure data
| Related structure data |  5sy1MC  5k8qC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8315.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8315.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of STRA6/CaM complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.255 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
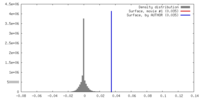
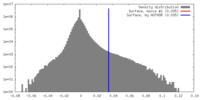
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_8315_msk_1.map emd_8315_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Reconstruction of STRA6/CaM complex, half map 1
| File | emd_8315_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of STRA6/CaM complex, half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Reconstruction of STRA6/CaM complex, half map 2
| File | emd_8315_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of STRA6/CaM complex, half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of zebrafish (D. rerio) STRA6 with copurified calmodulin ...
| Entire | Name: Complex of zebrafish (D. rerio) STRA6 with copurified calmodulin reconstituted in amphipol A8-35 |
|---|---|
| Components |
|
-Supramolecule #1: Complex of zebrafish (D. rerio) STRA6 with copurified calmodulin ...
| Supramolecule | Name: Complex of zebrafish (D. rerio) STRA6 with copurified calmodulin reconstituted in amphipol A8-35 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Calmodulin
| Macromolecule | Name: Calmodulin / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.82552 KDa |
| Sequence | String: MADQLTEEQI AEFKEAFSLF DKDGDGTITT KELGTVMRSL GQNPTEAELQ DMINEVDADG NGTIDFPEFL TMMARKMKDT DSEEEIREA FRVFDKDGNG FISAAELRHV MTNLGEKLTD EEVDEMIREA DIDGDGQVNY EEFVTMMTSK |
-Macromolecule #2: STRA6
| Macromolecule | Name: STRA6 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 75.551523 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSAETVNNYD YSDWYENAAP TKAPVEVIPP CDPTADEGLF HICIAAISLV VMLVLAILAR RQKLSDNQRG LTGLLSPVNF LDHTQHKGL AVAVYGVLFC KLVGMVLSHH PLPFTKEVAN KEFWMILALL YYPTLYYPLL ACGTLHNKVG YVLGSLLSWT H FGILVWQK ...String: MSAETVNNYD YSDWYENAAP TKAPVEVIPP CDPTADEGLF HICIAAISLV VMLVLAILAR RQKLSDNQRG LTGLLSPVNF LDHTQHKGL AVAVYGVLFC KLVGMVLSHH PLPFTKEVAN KEFWMILALL YYPTLYYPLL ACGTLHNKVG YVLGSLLSWT H FGILVWQK VDCPKTPQIY KYYALFGSLP QIACLAFLSF QYPLLLFKGL QNTETANASE DLSSSYYRDY VKKILKKKKP TK ISSSTSK PKLFDRLRDA VKSYIYTPED VFRFPLKLAI SVVVAFIALY QMALLLISGV LPTLHIVRRG VDENIAFLLA GFN IILSND RQEVVRIVVY YLWCVEICYV SAVTLSCLVN LLMLMRSMVL HRSNLKGLYR GDSLNVFNCH RSIRPSRPAL VCWM GFTSY QAAFLCLGMA IQTLVFFICI LFAVFLIIIP ILWGTNLMLF HIIGNLWPFW LTLVLAALIQ HVASRFLFIR KDGGT RDLN NRGSLFLLSY ILFLVNVMIG VVLGIWRVVI TALFNIVHLG RLDISLLNRN VEAFDPGYRC YSHYLKIEVS QSHPVM KAF CGLLLQSSGQ DGLSAQRIRD AEEGIQLVQQ EKKQNKVSNA KRARAHWQLL YTLVNNPSLV GSRKHFQCQS SESFING AL SRTSKEGSKK DGSVKEPNKE AESAAASN UniProtKB: Receptor for retinol uptake stra6 |
-Macromolecule #3: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 3 / Number of copies: 8 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #4: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 4 / Number of copies: 2 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.6 mg/mL |
|---|---|
| Buffer | pH: 7 |
| Grid | Model: Quantifoil, UltrAuFoil, R1.2/1.3 / Material: GOLD / Mesh: 400 / Pretreatment - Type: PLASMA CLEANING |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: 3 uL sample applied, 3-4 second blot time, 30 second wait time, blot force 3, grid blotted from both sides, plunged into liquid ethane (FEI VITROBOT MARK IV).. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 100.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-5sy1: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)