+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8298 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Iho670 Flagellar-like Filament | |||||||||
 Map data Map data | 3D cryo-EM reconstruction of the I. hospitalis Iho670 flagellar-like filament | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | immunoglobulin fold / Type IV pili / flagellin / CELL ADHESION | |||||||||
| Function / homology | Flagellin/pilin, N-terminal / membrane / Archaeal Type IV pilin N-terminal domain-containing protein Function and homology information Function and homology information | |||||||||
| Biological species |   Ignicoccus hospitalis (archaea) Ignicoccus hospitalis (archaea) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | Braun T / Vos M | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2016 Journal: Proc Natl Acad Sci U S A / Year: 2016Title: Archaeal flagellin combines a bacterial type IV pilin domain with an Ig-like domain. Authors: Tatjana Braun / Matthijn R Vos / Nir Kalisman / Nicholas E Sherman / Reinhard Rachel / Reinhard Wirth / Gunnar F Schröder / Edward H Egelman /     Abstract: The bacterial flagellar apparatus, which involves ∼40 different proteins, has been a model system for understanding motility and chemotaxis. The bacterial flagellar filament, largely composed of a ...The bacterial flagellar apparatus, which involves ∼40 different proteins, has been a model system for understanding motility and chemotaxis. The bacterial flagellar filament, largely composed of a single protein, flagellin, has been a model for understanding protein assembly. This system has no homology to the eukaryotic flagellum, in which the filament alone, composed of a microtubule-based axoneme, contains more than 400 different proteins. The archaeal flagellar system is simpler still, in some cases having ∼13 different proteins with a single flagellar filament protein. The archaeal flagellar system has no homology to the bacterial one and must have arisen by convergent evolution. However, it has been understood that the N-terminal domain of the archaeal flagellin is a homolog of the N-terminal domain of bacterial type IV pilin, showing once again how proteins can be repurposed in evolution for different functions. Using cryo-EM, we have been able to generate a nearly complete atomic model for a flagellar-like filament of the archaeon Ignicoccus hospitalis from a reconstruction at ∼4-Å resolution. We can now show that the archaeal flagellar filament contains a β-sandwich, previously seen in the FlaF protein that forms the anchor for the archaeal flagellar filament. In contrast to the bacterial flagellar filament, where the outer globular domains make no contact with each other and are not necessary for either assembly or motility, the archaeal flagellin outer domains make extensive contacts with each other that largely determine the interesting mechanical properties of these filaments, allowing these filaments to flex. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8298.map.gz emd_8298.map.gz | 48 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8298-v30.xml emd-8298-v30.xml emd-8298.xml emd-8298.xml | 9.8 KB 9.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8298.png emd_8298.png | 400.7 KB | ||
| Filedesc metadata |  emd-8298.cif.gz emd-8298.cif.gz | 4.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8298 http://ftp.pdbj.org/pub/emdb/structures/EMD-8298 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8298 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8298 | HTTPS FTP |
-Validation report
| Summary document |  emd_8298_validation.pdf.gz emd_8298_validation.pdf.gz | 552.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_8298_full_validation.pdf.gz emd_8298_full_validation.pdf.gz | 552.3 KB | Display | |
| Data in XML |  emd_8298_validation.xml.gz emd_8298_validation.xml.gz | 6.3 KB | Display | |
| Data in CIF |  emd_8298_validation.cif.gz emd_8298_validation.cif.gz | 7.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8298 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8298 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8298 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8298 | HTTPS FTP |
-Related structure data
| Related structure data |  5kyhMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8298.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8298.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D cryo-EM reconstruction of the I. hospitalis Iho670 flagellar-like filament | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
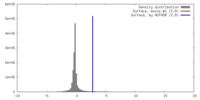
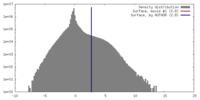
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Iho670 filament
| Entire | Name: Iho670 filament |
|---|---|
| Components |
|
-Supramolecule #1: Iho670 filament
| Supramolecule | Name: Iho670 filament / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Ignicoccus hospitalis (archaea) Ignicoccus hospitalis (archaea) |
-Macromolecule #1: Iho670
| Macromolecule | Name: Iho670 / type: protein_or_peptide / ID: 1 / Number of copies: 21 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Ignicoccus hospitalis (archaea) Ignicoccus hospitalis (archaea) |
| Molecular weight | Theoretical: 32.521539 KDa |
| Recombinant expression | Organism:   Ignicoccus hospitalis (archaea) Ignicoccus hospitalis (archaea) |
| Sequence | String: VSPVIATLLL ILIAVAAAVL LYTWVSGLSA NVAGTQVTGK SLTLIQATWA RPATNVGTTI SKDSFDRSKA VLILSFQPPA QVLQGGQAI TIDAIDVLYQ GRVVCHYDSF PMTADDKYHI GQTIGGLTAF GLVFWGFVGS TLSDFDAHNE TLVPGGIIHG K SDLATQPA ...String: VSPVIATLLL ILIAVAAAVL LYTWVSGLSA NVAGTQVTGK SLTLIQATWA RPATNVGTTI SKDSFDRSKA VLILSFQPPA QVLQGGQAI TIDAIDVLYQ GRVVCHYDSF PMTADDKYHI GQTIGGLTAF GLVFWGFVGS TLSDFDAHNE TLVPGGIIHG K SDLATQPA ARGYLGGDKG VTGEVHPGEK YKPDILLSFD DQYPFATILA GTWEVNYVST NYVETNFRNT SAVIKFDRFV NT HYSPDTQ NNNGVPIFDV ASASQSNFAV VIWCPNVNPN VMQSVDVKMV FSDGSTWEAS VPLSIT UniProtKB: Archaeal Type IV pilin N-terminal domain-containing protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 6 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 5.3 Å Applied symmetry - Helical parameters - Δ&Phi: 106.65 ° Applied symmetry - Helical parameters - Axial symmetry: C1 (asymmetric) Algorithm: BACK PROJECTION / Resolution.type: BY AUTHOR / Resolution: 4.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 146696 |
|---|---|
| Startup model | Type of model: OTHER / Details: solid cylinder |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)