[English] 日本語
 Yorodumi
Yorodumi- EMDB-7956: Negative stain EM map of Pan-EBOV antibody 515 Fab in complex wit... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7956 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Negative stain EM map of Pan-EBOV antibody 515 Fab in complex with EBOV GPdMuc trimer | |||||||||
 Map data Map data | Negative stain EM map of Pan-EBOV antibody 515 Fab in complex with EBOV GPdMuc. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 13.4 Å | |||||||||
 Authors Authors | Ward AB / Turner HL | |||||||||
 Citation Citation |  Journal: Immunity / Year: 2018 Journal: Immunity / Year: 2018Title: Multifunctional Pan-ebolavirus Antibody Recognizes a Site of Broad Vulnerability on the Ebolavirus Glycoprotein. Authors: Pavlo Gilchuk / Natalia Kuzmina / Philipp A Ilinykh / Kai Huang / Bronwyn M Gunn / Aubrey Bryan / Edgar Davidson / Benjamin J Doranz / Hannah L Turner / Marnie L Fusco / Matthew S Bramble / ...Authors: Pavlo Gilchuk / Natalia Kuzmina / Philipp A Ilinykh / Kai Huang / Bronwyn M Gunn / Aubrey Bryan / Edgar Davidson / Benjamin J Doranz / Hannah L Turner / Marnie L Fusco / Matthew S Bramble / Nicole A Hoff / Elad Binshtein / Nurgun Kose / Andrew I Flyak / Robin Flinko / Chiara Orlandi / Robert Carnahan / Erica H Parrish / Alexander M Sevy / Robin G Bombardi / Prashant K Singh / Patrick Mukadi / Jean Jacques Muyembe-Tamfum / Melanie D Ohi / Erica Ollmann Saphire / George K Lewis / Galit Alter / Andrew B Ward / Anne W Rimoin / Alexander Bukreyev / James E Crowe /   Abstract: Ebolaviruses cause severe disease in humans, and identification of monoclonal antibodies (mAbs) that are effective against multiple ebolaviruses are important for therapeutics development. Here we ...Ebolaviruses cause severe disease in humans, and identification of monoclonal antibodies (mAbs) that are effective against multiple ebolaviruses are important for therapeutics development. Here we describe a distinct class of broadly neutralizing human mAbs with protective capacity against three ebolaviruses infectious for humans: Ebola (EBOV), Sudan (SUDV), and Bundibugyo (BDBV) viruses. We isolated mAbs from human survivors of ebolavirus disease and identified a potent mAb, EBOV-520, which bound to an epitope in the glycoprotein (GP) base region. EBOV-520 efficiently neutralized EBOV, BDBV, and SUDV and also showed protective capacity in relevant animal models of these infections. EBOV-520 mediated protection principally by direct virus neutralization and exhibited multifunctional properties. This study identified a potent naturally occurring mAb and defined key features of the human antibody response that may contribute to broad protection. This multifunctional mAb and related clones are promising candidates for development as broadly protective pan-ebolavirus therapeutic molecules. | |||||||||
| History |
|
- Structure visualization
Structure visualization
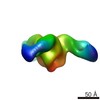
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7956.map.gz emd_7956.map.gz | 6.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7956-v30.xml emd-7956-v30.xml emd-7956.xml emd-7956.xml | 11.3 KB 11.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7956.png emd_7956.png | 49.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7956 http://ftp.pdbj.org/pub/emdb/structures/EMD-7956 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7956 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7956 | HTTPS FTP |
-Validation report
| Summary document |  emd_7956_validation.pdf.gz emd_7956_validation.pdf.gz | 77.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_7956_full_validation.pdf.gz emd_7956_full_validation.pdf.gz | 76.8 KB | Display | |
| Data in XML |  emd_7956_validation.xml.gz emd_7956_validation.xml.gz | 495 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7956 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7956 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7956 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7956 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_7956.map.gz / Format: CCP4 / Size: 11.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7956.map.gz / Format: CCP4 / Size: 11.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Negative stain EM map of Pan-EBOV antibody 515 Fab in complex with EBOV GPdMuc. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
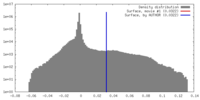
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : EBOV GPdMuc in complex with Pan-Ebolavirus antibody 515 Fab
| Entire | Name: EBOV GPdMuc in complex with Pan-Ebolavirus antibody 515 Fab |
|---|---|
| Components |
|
-Supramolecule #1: EBOV GPdMuc in complex with Pan-Ebolavirus antibody 515 Fab
| Supramolecule | Name: EBOV GPdMuc in complex with Pan-Ebolavirus antibody 515 Fab type: complex / ID: 1 / Parent: 0 / Details: Purified through SEC in TBS |
|---|
-Supramolecule #2: EBOV GPdMuc
| Supramolecule | Name: EBOV GPdMuc / type: complex / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Supramolecule #3: Pan-Ebolavirus antibody 515 Fab
| Supramolecule | Name: Pan-Ebolavirus antibody 515 Fab / type: complex / ID: 3 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Component - Name: TBS |
|---|---|
| Staining | Type: NEGATIVE / Material: Uranyl Formate |
| Grid | Material: COPPER / Support film - Material: CELLULOSE ACETATE / Support film - topology: HOLEY |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Number grids imaged: 1 / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C3 (3 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 13.4 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: RELION / Number images used: 7086 |
|---|---|
| Initial angle assignment | Type: ANGULAR RECONSTITUTION / Software - Name: RELION (ver. 2.1) |
| Final angle assignment | Type: ANGULAR RECONSTITUTION / Software - Name: RELION (ver. 2.1) |
| Final 3D classification | Software - Name: RELION |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)