[English] 日本語
 Yorodumi
Yorodumi- EMDB-7907: Two retinoschisin molecules interacting laterally forming a unit ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7907 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
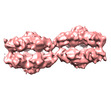
| Title | Two retinoschisin molecules interacting laterally forming a unit cell of a filamentous strand. | |||||||||
 Map data Map data | RS1 linear strand unit cell with two double octamer ring molecules. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Retinal adhesion protein X-linked retinoschisis Discoidin domain / CELL ADHESION | |||||||||
| Function / homology |  Function and homology information Function and homology informationneuron to neuron synapse / retina layer formation / eye development / phosphatidylinositol-3,4-bisphosphate binding / phosphatidylserine binding / photoreceptor inner segment / visual perception / protein homooligomerization / cell adhesion / external side of plasma membrane ...neuron to neuron synapse / retina layer formation / eye development / phosphatidylinositol-3,4-bisphosphate binding / phosphatidylserine binding / photoreceptor inner segment / visual perception / protein homooligomerization / cell adhesion / external side of plasma membrane / protein-containing complex binding / protein-containing complex / extracellular space Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
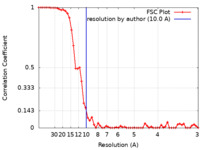
| Method | single particle reconstruction / cryo EM / Resolution: 10.0 Å | |||||||||
 Authors Authors | Heymann JB / Vijayasarathy C / Huang RK / Dearborn AD / Sieving PA / Steven AC | |||||||||
 Citation Citation |  Journal: J Cell Biol / Year: 2019 Journal: J Cell Biol / Year: 2019Title: Cryo-EM of retinoschisin branched networks suggests an intercellular adhesive scaffold in the retina. Authors: J Bernard Heymann / Camasamudram Vijayasarathy / Rick K Huang / Altaira D Dearborn / Paul A Sieving / Alasdair C Steven /  Abstract: Mutations in the retinal protein retinoschisin (RS1) cause progressive loss of vision in young males, a form of macular degeneration called X-linked retinoschisis (XLRS). We previously solved the ...Mutations in the retinal protein retinoschisin (RS1) cause progressive loss of vision in young males, a form of macular degeneration called X-linked retinoschisis (XLRS). We previously solved the structure of RS1, a 16-mer composed of paired back-to-back octameric rings. Here, we show by cryo-electron microscopy that RS1 16-mers can assemble into extensive branched networks. We classified the different configurations, finding four types of interaction between the RS1 molecules. The predominant configuration is a linear strand with a wavy appearance. Three less frequent types constitute the branch points of the network. In all cases, the "spikes" around the periphery of the double rings are involved in these interactions. In the linear strand, a loop (usually referred to as spike 1) occurs on both sides of the interface between neighboring molecules. Mutations in this loop suppress secretion, indicating the possibility of intracellular higher-order assembly. These observations suggest that branched networks of RS1 may play a stabilizing role in maintaining the integrity of the retina. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7907.map.gz emd_7907.map.gz | 37.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7907-v30.xml emd-7907-v30.xml emd-7907.xml emd-7907.xml | 17.7 KB 17.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7907_fsc.xml emd_7907_fsc.xml | 5.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_7907.png emd_7907.png | 93.6 KB | ||
| Masks |  emd_7907_msk_1.map emd_7907_msk_1.map | 42.9 MB |  Mask map Mask map | |
| Others |  emd_7907_half_map_1.map.gz emd_7907_half_map_1.map.gz emd_7907_half_map_2.map.gz emd_7907_half_map_2.map.gz | 39.5 MB 39.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7907 http://ftp.pdbj.org/pub/emdb/structures/EMD-7907 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7907 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7907 | HTTPS FTP |
-Validation report
| Summary document |  emd_7907_validation.pdf.gz emd_7907_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_7907_full_validation.pdf.gz emd_7907_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_7907_validation.xml.gz emd_7907_validation.xml.gz | 13.4 KB | Display | |
| Data in CIF |  emd_7907_validation.cif.gz emd_7907_validation.cif.gz | 17.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7907 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7907 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7907 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7907 | HTTPS FTP |
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7907.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7907.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RS1 linear strand unit cell with two double octamer ring molecules. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_7907_msk_1.map emd_7907_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Second half map of RS1 unit
| File | emd_7907_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Second half map of RS1 unit | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: First half map of the RS1 unit cell
| File | emd_7907_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | First half map of the RS1 unit cell | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Retinoschisin
| Entire | Name: Retinoschisin |
|---|---|
| Components |
|
-Supramolecule #1: Retinoschisin
| Supramolecule | Name: Retinoschisin / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Human retinoschisin with a honeybee mellitin leader sequence and a C-terminal 6-histidine tag |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Organ: Eye / Tissue: Retina / Location in cell: Intercellular space Homo sapiens (human) / Organ: Eye / Tissue: Retina / Location in cell: Intercellular space |
| Molecular weight | Theoretical: 380 KDa |
-Macromolecule #1: Retinoschisin
| Macromolecule | Name: Retinoschisin / type: protein_or_peptide / ID: 1 Details: Mature human RS1 with a C-terminal hexahistidine tag Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  unidentified baculovirus unidentified baculovirus |
| Sequence | String: STEDEGEDPW YQKACKCDCQ GGPNALWSAG ATSLDCIPEC PYHKPLGFES GEVTPDQITC SNPEQYVGWY SSWTANKARL NSQGFGCAWL SKFQDSSQWL QIDLKEIKVI SGILTQGRCD IDEWMTKYSV QYRTDERLNW IYYKDQTGNN RVFYGNS DR TSTVQNLLRP ...String: STEDEGEDPW YQKACKCDCQ GGPNALWSAG ATSLDCIPEC PYHKPLGFES GEVTPDQITC SNPEQYVGWY SSWTANKARL NSQGFGCAWL SKFQDSSQWL QIDLKEIKVI SGILTQGRCD IDEWMTKYSV QYRTDERLNW IYYKDQTGNN RVFYGNS DR TSTVQNLLRP PIISRFIRLI PLGWHVRIAI RMELLECVSK CAHHHHHH UniProtKB: Retinoschisin |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||
| Grid | Model: C-flat-2/2 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: PLASMA CLEANING | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 293 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 1-50 / Number grids imaged: 1 / Number real images: 1136 / Average exposure time: 0.25 sec. / Average electron dose: 0.66 e/Å2 / Details: 30 degree tilted specimen |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 3.23 µm / Calibrated defocus min: 1.04 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)