[English] 日本語
 Yorodumi
Yorodumi- EMDB-7875: BG505 MD39 SOSIP trimer in complex with mature BG18 fragment anti... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7875 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | BG505 MD39 SOSIP trimer in complex with mature BG18 fragment antigen binding | ||||||||||||
 Map data Map data | Sharpened map | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | HIV-1 / HIV Envelope / SOSIP / germline targeting / antibody / VIRAL PROTEIN / VIRAL PROTEIN-IMMUNE SYSTEM complex | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell ...symbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / identical protein binding / membrane Similarity search - Function | ||||||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
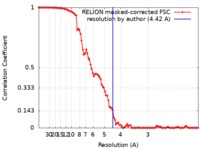
| Method | single particle reconstruction / cryo EM / Resolution: 4.42 Å | ||||||||||||
 Authors Authors | Ozorowski G / Steichen JM | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Science / Year: 2019 Journal: Science / Year: 2019Title: A generalized HIV vaccine design strategy for priming of broadly neutralizing antibody responses. Authors: Jon M Steichen / Ying-Cing Lin / Colin Havenar-Daughton / Simone Pecetta / Gabriel Ozorowski / Jordan R Willis / Laura Toy / Devin Sok / Alessia Liguori / Sven Kratochvil / Jonathan L Torres ...Authors: Jon M Steichen / Ying-Cing Lin / Colin Havenar-Daughton / Simone Pecetta / Gabriel Ozorowski / Jordan R Willis / Laura Toy / Devin Sok / Alessia Liguori / Sven Kratochvil / Jonathan L Torres / Oleksandr Kalyuzhniy / Eleonora Melzi / Daniel W Kulp / Sebastian Raemisch / Xiaozhen Hu / Steffen M Bernard / Erik Georgeson / Nicole Phelps / Yumiko Adachi / Michael Kubitz / Elise Landais / Jeffrey Umotoy / Amanda Robinson / Bryan Briney / Ian A Wilson / Dennis R Burton / Andrew B Ward / Shane Crotty / Facundo D Batista / William R Schief /  Abstract: Vaccine induction of broadly neutralizing antibodies (bnAbs) to HIV remains a major challenge. Germline-targeting immunogens hold promise for initiating the induction of certain bnAb classes; yet for ...Vaccine induction of broadly neutralizing antibodies (bnAbs) to HIV remains a major challenge. Germline-targeting immunogens hold promise for initiating the induction of certain bnAb classes; yet for most bnAbs, a strong dependence on antibody heavy chain complementarity-determining region 3 (HCDR3) is a major barrier. Exploiting ultradeep human antibody sequencing data, we identified a diverse set of potential antibody precursors for a bnAb with dominant HCDR3 contacts. We then developed HIV envelope trimer-based immunogens that primed responses from rare bnAb-precursor B cells in a mouse model and bound a range of potential bnAb-precursor human naïve B cells in ex vivo screens. Our repertoire-guided germline-targeting approach provides a framework for priming the induction of many HIV bnAbs and could be applied to most HCDR3-dominant antibodies from other pathogens. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7875.map.gz emd_7875.map.gz | 84.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7875-v30.xml emd-7875-v30.xml emd-7875.xml emd-7875.xml | 25.3 KB 25.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7875_fsc.xml emd_7875_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_7875.png emd_7875.png | 59.9 KB | ||
| Masks |  emd_7875_msk_1.map emd_7875_msk_1.map | 91.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-7875.cif.gz emd-7875.cif.gz | 7.6 KB | ||
| Others |  emd_7875_half_map_1.map.gz emd_7875_half_map_1.map.gz emd_7875_half_map_2.map.gz emd_7875_half_map_2.map.gz | 71 MB 71 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7875 http://ftp.pdbj.org/pub/emdb/structures/EMD-7875 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7875 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7875 | HTTPS FTP |
-Related structure data
| Related structure data |  6dfgMC  7876C  7884C  7885C  6dfhC  6nf5C  6nfcC  6oc7C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7875.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7875.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
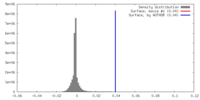
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_7875_msk_1.map emd_7875_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1
| File | emd_7875_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2
| File | emd_7875_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : BG505 MD39 SOSIP trimer in complex with mature BG18 fragment anti...
| Entire | Name: BG505 MD39 SOSIP trimer in complex with mature BG18 fragment antigen binding |
|---|---|
| Components |
|
-Supramolecule #1: BG505 MD39 SOSIP trimer in complex with mature BG18 fragment anti...
| Supramolecule | Name: BG505 MD39 SOSIP trimer in complex with mature BG18 fragment antigen binding type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Molecular weight | Theoretical: 570 KDa |
-Supramolecule #2: SOSIP trimer
| Supramolecule | Name: SOSIP trimer / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Supramolecule #3: BG18 fragment
| Supramolecule | Name: BG18 fragment / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Envelope glycoprotein gp160
| Macromolecule | Name: Envelope glycoprotein gp160 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 53.302312 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AENLWVTVYY GVPVWKDAET TLFCASDAKA YETEKHNVWA THACVPTDPN PQEIHLENVT EEFNMWKNNM VEQMHEDIIS LWDQSLKPC VKLTPLCVTL QCTNVTNNIT DDMRGELKNC SFNMTTELRD KKQKVYSLFY RLDVVQINEN QGNRSNNSNK E YRLINCNT ...String: AENLWVTVYY GVPVWKDAET TLFCASDAKA YETEKHNVWA THACVPTDPN PQEIHLENVT EEFNMWKNNM VEQMHEDIIS LWDQSLKPC VKLTPLCVTL QCTNVTNNIT DDMRGELKNC SFNMTTELRD KKQKVYSLFY RLDVVQINEN QGNRSNNSNK E YRLINCNT SAITQACPKV SFEPIPIHYC APAGFAILKC KDKKFNGTGP CPSVSTVQCT HGIKPVVSTQ LLLNGSLAEE EV IIRSENI TNNAKNILVQ LNTPVQINCT RPNNNTVKSI RIGPGQAFYY TGDIIGDIRQ AHCNVSKATW NETLGKVVKQ LRK HFGNNT IIRFAQSSGG DLEVTTHSFN CGGEFFYCNT SGLFNSTWIS NTSVQGSNST GSNDSITLPC RIKQIINMWQ RIGQ AMYAP PIQGVIRCVS NITGLILTRD GGSTNSTTET FRPGGGDMRD NWRSELYKYK VVKIEPLGVA PTRCKRRVVG R UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #2: Envelope glycoprotein gp160
| Macromolecule | Name: Envelope glycoprotein gp160 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 18.250541 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AVGIGAVSLG FLGAAGSTMG AASMTLTVQA RNLLSGIVQQ QSNLLRAPEP QQHLLKDTHW GIKQLQARVL AVEHYLRDQQ LLGIWGCSG KLICCTNVPW NSSWSNRNLS EIWDNMTWLQ WDKEISNYTQ IIYGLLEESQ NQQEKNEQDL LALDGTKHHH H HH UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #3: mature BG18 fragment antigen binding heavy chain
| Macromolecule | Name: mature BG18 fragment antigen binding heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.043186 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLRESGPG LVKPSETLSL SCTVSNDSRP SDHSWTWVRQ SPGKALEWIG DIHYNGATTY NPSLRSRVRI ELDQSIPRFS LKMTSMTAA DTGMYYCARN AIRIYGVVAL GEWFHYGMDV WGQGTAVTVS SASTKGPSVF PLAPSSKSTS GGTAALGCLV K DYFPEPVT ...String: QVQLRESGPG LVKPSETLSL SCTVSNDSRP SDHSWTWVRQ SPGKALEWIG DIHYNGATTY NPSLRSRVRI ELDQSIPRFS LKMTSMTAA DTGMYYCARN AIRIYGVVAL GEWFHYGMDV WGQGTAVTVS SASTKGPSVF PLAPSSKSTS GGTAALGCLV K DYFPEPVT VSWNSGALTS GVHTFPAVLQ SSGLYSLSSV VTVPSSSLGT QTYICNVNHK PSNTKVDKKV EPKSC |
-Macromolecule #4: mature BG18 fragment antigen binding light chain
| Macromolecule | Name: mature BG18 fragment antigen binding light chain / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.937352 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: SSELTQPPSV SVSPGQTARI TCSGAPLTSR FTYWYRQKPG QAPVLIISRS SQRSSGWSGR FSASWSGTTV TLTIRGVQAD DEADYYCQS SDTSDSYKMF GGGTKLTVLG QPKAAPSVTL FPPSSEELQA NKATLVCLIS DFYPGAVTVA WKADSSPVKA G VETTTPSK ...String: SSELTQPPSV SVSPGQTARI TCSGAPLTSR FTYWYRQKPG QAPVLIISRS SQRSSGWSGR FSASWSGTTV TLTIRGVQAD DEADYYCQS SDTSDSYKMF GGGTKLTVLG QPKAAPSVTL FPPSSEELQA NKATLVCLIS DFYPGAVTVA WKADSSPVKA G VETTTPSK QSNNKYAASS YLSLTPEQWK SHRSYSCQVT HEGSTVEKTV APTECS |
-Macromolecule #11: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 11 / Number of copies: 18 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.75 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 10 sec. / Pretreatment - Atmosphere: OTHER / Details: Gatan Model 950 Advanced Plasma System | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV / Details: 3.5 second blot time. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Number real images: 691 / Average exposure time: 12.0 sec. / Average electron dose: 62.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-6dfg: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)