+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7628 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Hemagglutinin with spot-to-plunge time of 100ms | |||||||||
 Map data Map data | Hemagglutinin with spot-to-plunge time of 100ms | |||||||||
 Sample Sample |
| |||||||||
| Biological species | unidentified (others) | |||||||||
| Method | electron tomography / cryo EM | |||||||||
 Authors Authors | Noble AJ / Wei H / Dandey VP / Zhang Z / Potter CS / Carragher B | |||||||||
 Citation Citation |  Journal: Nat Methods / Year: 2018 Journal: Nat Methods / Year: 2018Title: Reducing effects of particle adsorption to the air-water interface in cryo-EM. Authors: Alex J Noble / Hui Wei / Venkata P Dandey / Zhening Zhang / Yong Zi Tan / Clinton S Potter / Bridget Carragher /  Abstract: Most protein particles prepared in vitreous ice for single-particle cryo-electron microscopy (cryo-EM) are adsorbed to air-water or substrate-water interfaces, which can cause the particles to adopt ...Most protein particles prepared in vitreous ice for single-particle cryo-electron microscopy (cryo-EM) are adsorbed to air-water or substrate-water interfaces, which can cause the particles to adopt preferred orientations. By using a rapid plunge-freezing robot and nanowire grids, we were able to reduce some of the deleterious effects of the air-water interface by decreasing the dwell time of particles in thin liquid films. We demonstrated this by using single-particle cryo-EM and cryo-electron tomography (cryo-ET) to examine hemagglutinin, insulin receptor complex, and apoferritin. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7628.map.gz emd_7628.map.gz | 1.6 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7628-v30.xml emd-7628-v30.xml emd-7628.xml emd-7628.xml | 8 KB 8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7628.png emd_7628.png | 428.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7628 http://ftp.pdbj.org/pub/emdb/structures/EMD-7628 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7628 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7628 | HTTPS FTP |
-Related structure data
| Related structure data |  7623C  7624C  7625C  7627C  7629C  7630C  7788C  7791C  7792C C: citing same article ( |
|---|---|
| EM raw data |  EMPIAR-10172 (Title: CryoET of hemagglutinin single particle with spot-to-plunge time of 100ms EMPIAR-10172 (Title: CryoET of hemagglutinin single particle with spot-to-plunge time of 100msData size: 21.9 / Data #1: Tilt-series image stacks [tilt series] Data #2: Appion-Protomo tilt-series alignments [tilt series]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7628.map.gz / Format: CCP4 / Size: 1.9 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7628.map.gz / Format: CCP4 / Size: 1.9 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Hemagglutinin with spot-to-plunge time of 100ms | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.34 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
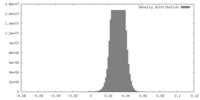
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
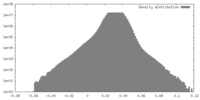
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Hemagglutinin
| Entire | Name: Hemagglutinin |
|---|---|
| Components |
|
-Supramolecule #1: Hemagglutinin
| Supramolecule | Name: Hemagglutinin / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism: unidentified (others) |
| Recombinant expression | Organism: unidentified (others) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 0.0001 |
|---|---|
| Grid | Model: Homemade |
| Vitrification | Cryogen name: ETHANE / Instrument: OTHER Details: The value given for _emd_vitrification.instrument is SPOTITON. This is not in a list of allowed values set(['LEICA EM CPC', 'GATAN CRYOPLUNGE 3', 'LEICA PLUNGER', 'FEI VITROBOT MARK II', ...Details: The value given for _emd_vitrification.instrument is SPOTITON. This is not in a list of allowed values set(['LEICA EM CPC', 'GATAN CRYOPLUNGE 3', 'LEICA PLUNGER', 'FEI VITROBOT MARK II', 'HOMEMADE PLUNGER', 'REICHERT-JUNG PLUNGER', 'FEI VITROBOT MARK I', 'LEICA KF80', 'FEI VITROBOT MARK III', 'LEICA EM GP', 'OTHER', 'FEI VITROBOT MARK IV']) so OTHER is written into the XML file. |
| Sectioning | Other: NO SECTIONING |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: DIRECT ELECTRON DE-20 (5k x 3k) / Detector mode: INTEGRATING / Average electron dose: 3.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Algorithm: SIMULTANEOUS ITERATIVE (SIRT) / Software - Name: TOMO3D / Software - details: SIRT, 30 iterations / Number images used: 30 |
|---|
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)