[English] 日本語
 Yorodumi
Yorodumi- EMDB-7588: Promotion of Virus Assembly and Organization by the Measles Virus... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7588 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Promotion of Virus Assembly and Organization by the Measles Virus Matrix Protein: F glycoprotein trimer | |||||||||||||||||||||
 Map data Map data | Measles virus fusion glycoprotein trimer from rec-MeV infected cells | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
| Biological species |  | |||||||||||||||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 20.0 Å | |||||||||||||||||||||
 Authors Authors | Ke Z / Strauss JD / Hampton CM / Dillard RS / Wright ER | |||||||||||||||||||||
| Funding support |  United States, 6 items United States, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2018 Journal: Nat Commun / Year: 2018Title: Promotion of virus assembly and organization by the measles virus matrix protein. Authors: Zunlong Ke / Joshua D Strauss / Cheri M Hampton / Melinda A Brindley / Rebecca S Dillard / Fredrick Leon / Kristen M Lamb / Richard K Plemper / Elizabeth R Wright /  Abstract: Measles virus (MeV) remains a major human pathogen, but there are presently no licensed antivirals to treat MeV or other paramyxoviruses. Here, we use cryo-electron tomography (cryo-ET) to elucidate ...Measles virus (MeV) remains a major human pathogen, but there are presently no licensed antivirals to treat MeV or other paramyxoviruses. Here, we use cryo-electron tomography (cryo-ET) to elucidate the principles governing paramyxovirus assembly in MeV-infected human cells. The three-dimensional (3D) arrangement of the MeV structural proteins including the surface glycoproteins (F and H), matrix protein (M), and the ribonucleoprotein complex (RNP) are characterized at stages of virus assembly and budding, and in released virus particles. The M protein is observed as an organized two-dimensional (2D) paracrystalline array associated with the membrane. A two-layered F-M lattice is revealed suggesting that interactions between F and M may coordinate processes essential for MeV assembly. The RNP complex remains associated with and in close proximity to the M lattice. In this model, the M lattice facilitates the well-ordered incorporation and concentration of the surface glycoproteins and the RNP at sites of virus assembly. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7588.map.gz emd_7588.map.gz | 949.7 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7588-v30.xml emd-7588-v30.xml emd-7588.xml emd-7588.xml | 14.3 KB 14.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7588.png emd_7588.png | 18.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7588 http://ftp.pdbj.org/pub/emdb/structures/EMD-7588 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7588 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7588 | HTTPS FTP |
-Related structure data
| Related structure data |  7565C  7566C  7587C  7590C  7591C  7594C  7595C  7596C  7597C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7588.map.gz / Format: CCP4 / Size: 1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7588.map.gz / Format: CCP4 / Size: 1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Measles virus fusion glycoprotein trimer from rec-MeV infected cells | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 5.88 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
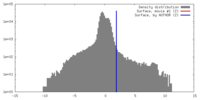
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Measles virus
| Entire | Name:  |
|---|---|
| Components |
|
-Supramolecule #1: Measles virus
| Supramolecule | Name: Measles virus / type: virus / ID: 1 / Parent: 0 / Details: Measles virus F glycoprotein trimer / NCBI-ID: 11234 / Sci species name: Measles virus / Sci species strain: recMeV / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: GOLD / Mesh: 200 / Support film - Material: CARBON |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 85 % / Chamber temperature: 293 K / Instrument: GATAN CRYOPLUNGE 3 |
| Details | measles virus infected cells |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2200FS |
|---|---|
| Specialist optics | Energy filter - Name: In-column Omega Filter / Energy filter - Lower energy threshold: 0 eV / Energy filter - Upper energy threshold: 20 eV |
| Image recording | Film or detector model: DIRECT ELECTRON DE-20 (5k x 3k) / Digitization - Dimensions - Width: 5080 pixel / Digitization - Dimensions - Height: 3768 pixel / Digitization - Frames/image: 1-8 / Average exposure time: 0.4 sec. / Average electron dose: 2.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 8.0 µm / Nominal defocus min: 4.0 µm / Nominal magnification: 20000 |
| Sample stage | Specimen holder model: GATAN 914 HIGH TILT LIQUID NITROGEN CRYO TRANSFER TOMOGRAPHY HOLDER Cooling holder cryogen: NITROGEN |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C3 (3 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 20.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: PEET (ver. 1.11.0 alpha) / Number subtomograms used: 539 |
|---|---|
| Extraction | Number tomograms: 1 / Number images used: 539 / Method: volumes picked manually in IMOD slicer window / Software - Name: PEET (ver. 1.11.0 alpha) |
| CTF correction | Software - Name: eTomo (ver. 4.9.4) |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)