[English] 日本語
 Yorodumi
Yorodumi- EMDB-7559: Bacteriophage A511 baseplate in post-host attachment state (urea-... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7559 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Bacteriophage A511 baseplate in post-host attachment state (urea-induced) | |||||||||
 Map data Map data | Bacteriophage A511 in post-host attachment contracted state (urea-induced) | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Listeria virus A511 Listeria virus A511 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 14.0 Å | |||||||||
 Authors Authors | Guerrero-Ferreira R | |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2019 Journal: EMBO J / Year: 2019Title: Structure and transformation of bacteriophage A511 baseplate and tail upon infection of cells. Authors: Ricardo C Guerrero-Ferreira / Mario Hupfeld / Sergey Nazarov / Nicholas Mi Taylor / Mikhail M Shneider / Jagan M Obbineni / Martin J Loessner / Takashi Ishikawa / Jochen Klumpp / Petr G Leiman /    Abstract: Contractile injection systems (bacteriophage tails, type VI secretions system, R-type pyocins, etc.) utilize a rigid tube/contractile sheath assembly for breaching the envelope of bacterial and ...Contractile injection systems (bacteriophage tails, type VI secretions system, R-type pyocins, etc.) utilize a rigid tube/contractile sheath assembly for breaching the envelope of bacterial and eukaryotic cells. Among contractile injection systems, bacteriophages that infect Gram-positive bacteria represent the least understood members. Here, we describe the structure of bacteriophage A511 tail in its pre- and post-host attachment states (extended and contracted, respectively) using cryo-electron microscopy, cryo-electron tomography, and X-ray crystallography. We show that the structure of the tube-baseplate complex of A511 is similar to that of phage T4, but the A511 baseplate is decorated with different receptor-binding proteins, which undergo a large structural transformation upon host attachment and switch the symmetry of the baseplate-tail fiber assembly from threefold to sixfold. For the first time under native conditions, we show that contraction of the phage tail sheath assembly starts at the baseplate and propagates through the sheath in a domino-like motion. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7559.map.gz emd_7559.map.gz | 96.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7559-v30.xml emd-7559-v30.xml emd-7559.xml emd-7559.xml | 11.4 KB 11.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7559_fsc.xml emd_7559_fsc.xml | 10 KB | Display |  FSC data file FSC data file |
| Images |  emd_7559.png emd_7559.png | 60.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7559 http://ftp.pdbj.org/pub/emdb/structures/EMD-7559 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7559 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7559 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_7559.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7559.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Bacteriophage A511 in post-host attachment contracted state (urea-induced) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.27 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
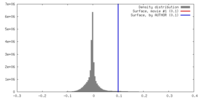
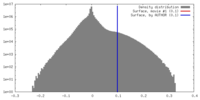
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Listeria virus A511
| Entire | Name:  Listeria virus A511 Listeria virus A511 |
|---|---|
| Components |
|
-Supramolecule #1: Listeria virus A511
| Supramolecule | Name: Listeria virus A511 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Propagated in Listeria ivanovii strain WSLC 3009 (SV 5) NCBI-ID: 40523 / Sci species name: Listeria virus A511 / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Listeria (bacteria) Listeria (bacteria) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Details: SM buffer: 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 8 mM MgSO4 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV |
| Details | Specimen was dialyzed into SM buffer containing 2M urea prior to cryo-EM sample preparation |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: FEI EAGLE (4k x 4k) / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: GATAN 626 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)