+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7482 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the SthK cyclic nucleotide-gated potassium channel | |||||||||
 Map data Map data | SthK ion channel apo structure, primary map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ion channel / tetramer / lipid / cyclic nucleotide / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationintracellularly cyclic nucleotide-activated monoatomic cation channel activity / protein-containing complex binding / membrane Similarity search - Function | |||||||||
| Biological species |  Spirochaeta thermophila DSM 6578 (bacteria) / Spirochaeta thermophila DSM 6578 (bacteria) /  Spirochaeta thermophila (bacteria) Spirochaeta thermophila (bacteria) | |||||||||
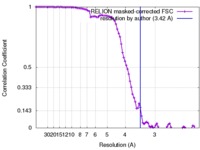
| Method | single particle reconstruction / cryo EM / Resolution: 3.42 Å | |||||||||
 Authors Authors | Nimigean CM / Rheinberger J | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2018 Journal: Elife / Year: 2018Title: Ligand discrimination and gating in cyclic nucleotide-gated ion channels from apo and partial agonist-bound cryo-EM structures. Authors: Jan Rheinberger / Xiaolong Gao / Philipp Am Schmidpeter / Crina M Nimigean /  Abstract: Cyclic nucleotide-modulated channels have important roles in visual signal transduction and pacemaking. Binding of cyclic nucleotides (cAMP/cGMP) elicits diverse functional responses in different ...Cyclic nucleotide-modulated channels have important roles in visual signal transduction and pacemaking. Binding of cyclic nucleotides (cAMP/cGMP) elicits diverse functional responses in different channels within the family despite their high sequence and structure homology. The molecular mechanisms responsible for ligand discrimination and gating are unknown due to lack of correspondence between structural information and functional states. Using single particle cryo-electron microscopy and single-channel recording, we assigned functional states to high-resolution structures of SthK, a prokaryotic cyclic nucleotide-gated channel. The structures for apo, cAMP-bound, and cGMP-bound SthK in lipid nanodiscs, correspond to no, moderate, and low single-channel activity, respectively, consistent with the observation that all structures are in resting, closed states. The similarity between apo and ligand-bound structures indicates that ligand-binding domains are strongly coupled to pore and SthK gates in an allosteric, concerted fashion. The different orientations of cAMP and cGMP in the 'resting' and 'activated' structures suggest a mechanism for ligand discrimination. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7482.map.gz emd_7482.map.gz | 59.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7482-v30.xml emd-7482-v30.xml emd-7482.xml emd-7482.xml | 12.6 KB 12.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7482_fsc.xml emd_7482_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_7482.png emd_7482.png | 86.4 KB | ||
| Filedesc metadata |  emd-7482.cif.gz emd-7482.cif.gz | 5.5 KB | ||
| Others |  emd_7482_additional.map.gz emd_7482_additional.map.gz | 60 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7482 http://ftp.pdbj.org/pub/emdb/structures/EMD-7482 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7482 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7482 | HTTPS FTP |
-Validation report
| Summary document |  emd_7482_validation.pdf.gz emd_7482_validation.pdf.gz | 594.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_7482_full_validation.pdf.gz emd_7482_full_validation.pdf.gz | 594.2 KB | Display | |
| Data in XML |  emd_7482_validation.xml.gz emd_7482_validation.xml.gz | 10.8 KB | Display | |
| Data in CIF |  emd_7482_validation.cif.gz emd_7482_validation.cif.gz | 14.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7482 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7482 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7482 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7482 | HTTPS FTP |
-Related structure data
| Related structure data |  6cjqMC  7483C  7484C  6cjtC  6cjuC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7482.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7482.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SthK ion channel apo structure, primary map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0961 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
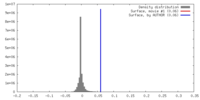
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: SthK ion channel apo structure, additional volume
| File | emd_7482_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SthK ion channel apo structure, additional volume | ||||||||||||
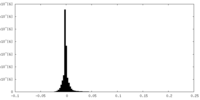
| Projections & Slices |
| ||||||||||||
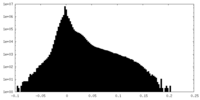
| Density Histograms |
- Sample components
Sample components
-Entire : SthK cyclic nucleotide-gated potassium channel
| Entire | Name: SthK cyclic nucleotide-gated potassium channel |
|---|---|
| Components |
|
-Supramolecule #1: SthK cyclic nucleotide-gated potassium channel
| Supramolecule | Name: SthK cyclic nucleotide-gated potassium channel / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Spirochaeta thermophila DSM 6578 (bacteria) Spirochaeta thermophila DSM 6578 (bacteria) |
-Macromolecule #1: SthK cyclic nucleotide-gated potassium channel
| Macromolecule | Name: SthK cyclic nucleotide-gated potassium channel / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Spirochaeta thermophila (bacteria) / Strain: ATCC 700085 / DSM 6578 / Z-1203 Spirochaeta thermophila (bacteria) / Strain: ATCC 700085 / DSM 6578 / Z-1203 |
| Molecular weight | Theoretical: 51.118574 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAKDIGINSD PNSSSVDKLM KSSGVSNPTY TLVWKVWILA VTLYYAIRIP LTLVFPSLFS PLLPLDILAS LALIADIPLD LAFESRRTS GRKPTLLAPS RLPDLLAALP LDLLVFALHL PSPLSLLSLV RLLKLISVQR SATRILSYRI NPALLRLLSL V GFILLAAH ...String: MAKDIGINSD PNSSSVDKLM KSSGVSNPTY TLVWKVWILA VTLYYAIRIP LTLVFPSLFS PLLPLDILAS LALIADIPLD LAFESRRTS GRKPTLLAPS RLPDLLAALP LDLLVFALHL PSPLSLLSLV RLLKLISVQR SATRILSYRI NPALLRLLSL V GFILLAAH GIACGWMSLQ PPSENPAGTR YLSAFYWTIT TLTTIGYGDI TPSTPTQTVY TIVIELLGAA MYGLVIGNIA SL VSKLDAA KLLHRERVER VTAFLSYKRI SPELQRRIIE YFDYLWETRR GYEEREVLKE LPHPLRLAVA MEIHGDVIEK VPL FKGAGE EFIRDIILHL EPVIYGPGEY IIRAGEMGSD VYFINRGSVE VLSADEKTRY AILSEGQFFG EMALILRAPR TATV RARAF CDLYRLDKET FDRILSRYPE IAAQIQELAV RRKELESSGL VPRGSVKHHH H UniProtKB: Transcriptional regulator, Crp/Fnr family |
-Macromolecule #2: (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]o...
| Macromolecule | Name: (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate type: ligand / ID: 2 / Number of copies: 24 / Formula: PGW |
|---|---|
| Molecular weight | Theoretical: 749.007 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)