[English] 日本語
 Yorodumi
Yorodumi- EMDB-7286: Cryo-EM density map of Alternative Complex III from Flavobacteriu... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7286 | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM density map of Alternative Complex III from Flavobacterium johnsoniae (Wild Type) | |||||||||||||||||||||||||||
 Map data Map data | Structure of Alternative Complex III from Flavobacterium johnsoniae (Wild Type), primary map | |||||||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||||||||
 Keywords Keywords | Electron transport chain / triacylated cysteine / heme c domain / iron-sulfur cluster / MEMBRANE PROTEIN | |||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationelectron transfer activity / heme binding / metal ion binding / membrane / plasma membrane Similarity search - Function | |||||||||||||||||||||||||||
| Biological species |  Flavobacterium johnsoniae UW101 (bacteria) Flavobacterium johnsoniae UW101 (bacteria) | |||||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||||||||||||||||||||
 Authors Authors | Benlekbir S / Rubinstein JL | |||||||||||||||||||||||||||
| Funding support |  United States, United States,  Canada, 8 items Canada, 8 items
| |||||||||||||||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2018 Journal: Nature / Year: 2018Title: Structure of the alternative complex III in a supercomplex with cytochrome oxidase. Authors: Chang Sun / Samir Benlekbir / Padmaja Venkatakrishnan / Yuhang Wang / Sangjin Hong / Jonathan Hosler / Emad Tajkhorshid / John L Rubinstein / Robert B Gennis /   Abstract: Alternative complex III (ACIII) is a key component of the respiratory and/or photosynthetic electron transport chains of many bacteria. Like complex III (also known as the bc complex), ACIII ...Alternative complex III (ACIII) is a key component of the respiratory and/or photosynthetic electron transport chains of many bacteria. Like complex III (also known as the bc complex), ACIII catalyses the oxidation of membrane-bound quinol and the reduction of cytochrome c or an equivalent electron carrier. However, the two complexes have no structural similarity. Although ACIII has eluded structural characterization, several of its subunits are known to be homologous to members of the complex iron-sulfur molybdoenzyme (CISM) superfamily , including the proton pump polysulfide reductase. We isolated the ACIII from Flavobacterium johnsoniae with native lipids using styrene maleic acid copolymer, both as an independent enzyme and as a functional 1:1 supercomplex with an aa-type cytochrome c oxidase (cyt aa). We determined the structure of ACIII to 3.4 Å resolution by cryo-electron microscopy and constructed an atomic model for its six subunits. The structure, which contains a [3Fe-4S] cluster, a [4Fe-4S] cluster and six haem c units, shows that ACIII uses known elements from other electron transport complexes arranged in a previously unknown manner. Modelling of the cyt aa component of the supercomplex revealed that it is structurally modified to facilitate association with ACIII, illustrating the importance of the supercomplex in this electron transport chain. The structure also resolves two of the subunits of ACIII that are anchored to the lipid bilayer with N-terminal triacylated cysteine residues, an important post-translational modification found in numerous prokaryotic membrane proteins that has not previously been observed structurally in a lipid bilayer. | |||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7286.map.gz emd_7286.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7286-v30.xml emd-7286-v30.xml emd-7286.xml emd-7286.xml | 25.5 KB 25.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7286.png emd_7286.png | 133.1 KB | ||
| Filedesc metadata |  emd-7286.cif.gz emd-7286.cif.gz | 7.7 KB | ||
| Others |  emd_7286_additional_1.map.gz emd_7286_additional_1.map.gz emd_7286_additional_2.map.gz emd_7286_additional_2.map.gz | 59.6 MB 59.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7286 http://ftp.pdbj.org/pub/emdb/structures/EMD-7286 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7286 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7286 | HTTPS FTP |
-Related structure data
| Related structure data |  6btmMC  7447C  7448C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7286.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7286.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of Alternative Complex III from Flavobacterium johnsoniae (Wild Type), primary map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
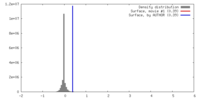
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: 3.6-angstrom map for the Alternative Complex III: cyt...
| File | emd_7286_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3.6-angstrom map for the Alternative Complex III: cyt aa3 oxidase supercomplex from Flavobacterium johnsoniae | ||||||||||||
| Projections & Slices |
| ||||||||||||
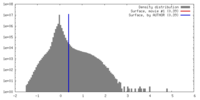
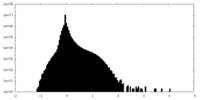
| Density Histograms |
-Additional map: 3.6-angstrom map for the Alternative Complex III from...
| File | emd_7286_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3.6-angstrom map for the Alternative Complex III from Flavobacterium johnsoniae | ||||||||||||
| Projections & Slices |
| ||||||||||||
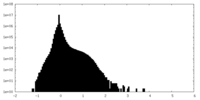
| Density Histograms |
- Sample components
Sample components
+Entire : ACIII-cyt aa3 supercomplex
+Supramolecule #1: ACIII-cyt aa3 supercomplex
+Macromolecule #1: Alternative Complex III subunit A
+Macromolecule #2: Alternative Complex III subunit B
+Macromolecule #3: Alternative Complex III subunit C
+Macromolecule #4: Alternative Complex III subunit D
+Macromolecule #5: Alternative Complex III subunit E
+Macromolecule #6: Alternative Complex III subunit F
+Macromolecule #7: HEME C
+Macromolecule #8: FE3-S4 CLUSTER
+Macromolecule #9: IRON/SULFUR CLUSTER
+Macromolecule #10: DECANOIC ACID
+Macromolecule #11: (2S)-3-hydroxypropane-1,2-diyl ditetradecanoate
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Material: GOLD |
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-40 / Number grids imaged: 2 / Number real images: 2017 / Average exposure time: 10.0 sec. / Average electron dose: 61.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER |
|---|---|
| Output model |  PDB-6btm: |
 Movie
Movie Controller
Controller




























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)