[English] 日本語
 Yorodumi
Yorodumi- EMDB-7269: Single-Molecule 3D Image of DNA Origami Bennett Linkage by Indivi... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7269 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Single-Molecule 3D Image of DNA Origami Bennett Linkage by Individual Particle Electron Tomography (No. 072) | |||||||||||||||
 Map data Map data | Origami Bennett Linkage (No. 072) | |||||||||||||||
 Sample Sample |
| |||||||||||||||
| Biological species |  Escherichia virus M13 / synthetic construct (others) Escherichia virus M13 / synthetic construct (others) | |||||||||||||||
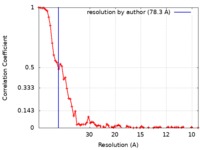
| Method | electron tomography / cryo EM / negative staining / Resolution: 78.3 Å | |||||||||||||||
 Authors Authors | Lei D / Marras A / Liu J / Huang C / Zhou L / Castro C / Su H / Ren G | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2018 Journal: Nat Commun / Year: 2018Title: Three-dimensional structural dynamics of DNA origami Bennett linkages using individual-particle electron tomography. Authors: Dongsheng Lei / Alexander E Marras / Jianfang Liu / Chao-Min Huang / Lifeng Zhou / Carlos E Castro / Hai-Jun Su / Gang Ren /  Abstract: Scaffolded DNA origami has proven to be a powerful and efficient technique to fabricate functional nanomachines by programming the folding of a single-stranded DNA template strand into three- ...Scaffolded DNA origami has proven to be a powerful and efficient technique to fabricate functional nanomachines by programming the folding of a single-stranded DNA template strand into three-dimensional (3D) nanostructures, designed to be precisely motion-controlled. Although two-dimensional (2D) imaging of DNA nanomachines using transmission electron microscopy and atomic force microscopy suggested these nanomachines are dynamic in 3D, geometric analysis based on 2D imaging was insufficient to uncover the exact motion in 3D. Here we use the individual-particle electron tomography method and reconstruct 129 density maps from 129 individual DNA origami Bennett linkage mechanisms at ~ 6-14 nm resolution. The statistical analyses of these conformations lead to understanding the 3D structural dynamics of Bennett linkage mechanisms. Moreover, our effort provides experimental verification of a theoretical kinematics model of DNA origami, which can be used as feedback to improve the design and control of motion via optimized DNA sequences and routing. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7269.map.gz emd_7269.map.gz | 24.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7269-v30.xml emd-7269-v30.xml emd-7269.xml emd-7269.xml | 11.7 KB 11.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7269_fsc.xml emd_7269_fsc.xml | 6.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_7269.png emd_7269.png | 27.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7269 http://ftp.pdbj.org/pub/emdb/structures/EMD-7269 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7269 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7269 | HTTPS FTP |
-Validation report
| Summary document |  emd_7269_validation.pdf.gz emd_7269_validation.pdf.gz | 78.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_7269_full_validation.pdf.gz emd_7269_full_validation.pdf.gz | 77.3 KB | Display | |
| Data in XML |  emd_7269_validation.xml.gz emd_7269_validation.xml.gz | 495 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7269 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7269 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7269 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7269 | HTTPS FTP |
-Related structure data
| Related structure data |  7155C  7156C  7157C  7158C  7159C  7160C  7161C  7162C  7163C  7164C  7165C  7166C  7167C  7168C  7169C  7170C  7171C  7172C  7173C  7174C  7175C  7176C  7177C  7178C  7179C  7180C  7181C  7182C  7183C  7184C  7185C  7186C  7187C  7188C  7189C  7190C  7191C  7192C  7193C  7194C  7195C  7196C  7197C  7198C  7199C  7200C  7201C  7202C  7203C  7204C  7205C  7206C  7207C  7208C  7209C  7210C  7211C  7212C  7213C  7214C  7215C  7216C  7217C  7218C  7219C  7220C  7221C  7222C  7223C  7224C  7225C  7226C  7227C  7228C  7229C  7230C  7231C  7232C  7233C  7234C  7235C  7236C  7237C  7238C  7239C  7240C  7241C  7242C  7243C  7244C  7245C  7246C  7247C  7248C  7249C  7250C  7251C  7252C  7253C  7254C  7255C  7256C  7257C  7258C  7259C  7260C  7261C  7262C  7263C  7264C  7265C  7266C  7267C  7268C  7270C  7271C  7272C  7273C  7274C  7275C  7276C  7277C  7278C  7279C  7280C  7281C  7282C  7283C  7284C  7285C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7269.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7269.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Origami Bennett Linkage (No. 072) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.8 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : DNA origami Bennett linkage
| Entire | Name: DNA origami Bennett linkage |
|---|---|
| Components |
|
-Supramolecule #1: DNA origami Bennett linkage
| Supramolecule | Name: DNA origami Bennett linkage / type: complex / ID: 1 / Parent: 0 |
|---|
-Supramolecule #2: M13 phage genome segment
| Supramolecule | Name: M13 phage genome segment / type: complex / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  Escherichia virus M13 Escherichia virus M13 |
| Recombinant expression | Organism:  |
-Supramolecule #3: Synthetic DNA oligonucleotides
| Supramolecule | Name: Synthetic DNA oligonucleotides / type: complex / ID: 3 / Parent: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Details: Tris-Borate-EDTA (TBE) buffer containing 11 mM MgCl2 |
|---|---|
| Staining | Type: NEGATIVE / Material: Uranyl Formate Details: The grid was washed twice with 1% (w/v) uranyl formate. |
| Grid | Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: LACEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.039 kPa |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 277 K / Instrument: LEICA EM GP |
| Details | DNA origami Bennett linkage was diluted to ~2 nM with Tris-Borate-EDTA (TBE) buffer containing 11 mM MgCl2. |
| Sectioning | Other: NO SECTIONING |
- Electron microscopy
Electron microscopy
| Microscope | ZEISS LIBRA120PLUS |
|---|---|
| Specialist optics | Energy filter - Name: In-column Omega Filter |
| Image recording | Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Average electron dose: 1.95 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.2 mm / Nominal magnification: 50000 |
| Sample stage | Specimen holder model: GATAN CT3500 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera

 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)