+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Trm10-tRNA complex (Two Trm10 monomers bound to one tRNA) | |||||||||
 Map data Map data | Full map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SPOUT methyltransferase / tRNA / methylation / RNA BINDING PROTEIN / RNA BINDING PROTEIN-RNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationtRNA (guanine) methyltransferase activity / tRNA (guanine9-N1)-methyltransferase / tRNA (guanosine(9)-N1)-methyltransferase activity / tRNA N1-guanine methylation / tRNA modification / tRNA methylation / tRNA binding / nucleoplasm / nucleus / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.89 Å | |||||||||
 Authors Authors | Nandi S / Strassler SE / Conn GL | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: bioRxiv / Year: 2025 Journal: bioRxiv / Year: 2025Title: Molecular basis of tRNA substrate recognition and modification by the atypical SPOUT methyltransferase Trm10. Authors: Suparno Nandi / Sarah E Strassler / Debayan Dey / Aiswarya Krishnamohan / George M Harris / Lindsay R Comstock / Jane E Jackman / Graeme L Conn /  Abstract: The evolutionarily conserved methyltransferase Trm10 modifies the N1 position of guanosine 9 (G9) in some tRNAs, but how the enzyme recognizes and modifies its substrate tRNAs remains unclear. Here, ...The evolutionarily conserved methyltransferase Trm10 modifies the N1 position of guanosine 9 (G9) in some tRNAs, but how the enzyme recognizes and modifies its substrate tRNAs remains unclear. Here, we used an S-adenosyl-L-methionine (SAM) analog to trap the Trm10-tRNA complex and enable determination of its structure in a post-catalytic state by cryogenic electron microscopy (cryo-EM). We observed three distinct complexes: two with a single Trm10 bound to tRNA that differ in their tRNA acceptor stem orientation ("closed" and "open") and a minor population with two Trm10s engaging the same tRNA. The monomeric complexes reveal a positively charged surface that guides the G9 into the catalytic site with key conserved residues forming "pincer"-like interactions that stabilize the flipped methylated nucleotide. In the open tRNA conformation, the acceptor stem is rotated away from the enzyme, weakening the tRNA-protein contacts, consistent with a product-release conformation. The dimeric complex, which is supported by tRNA-dependent protein crosslinking, reveals one Trm10 positioned similarly to the monomeric complexes and engaged with G9, while the other Trm10 contacts distal tRNA regions, suggesting a potential role in facilitating a key conformational transition during the process of catalysis or modified tRNA release. Finally, molecular dynamics simulations comparing G9- and A9-containing complexes reveal that G9 is efficiently stabilized in the binding pocket unlike A9, identifying the structural basis for guanosine selectivity. Overall, these findings reveal the structural determinants of G9-specific tRNA methylation by Trm10 and suggest a unique mechanism of action among RNA-modifying SPOUT methyltransferases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_72370.map.gz emd_72370.map.gz | 31.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-72370-v30.xml emd-72370-v30.xml emd-72370.xml emd-72370.xml | 23 KB 23 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_72370_fsc.xml emd_72370_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_72370.png emd_72370.png | 48.6 KB | ||
| Masks |  emd_72370_msk_1.map emd_72370_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-72370.cif.gz emd-72370.cif.gz | 6.9 KB | ||
| Others |  emd_72370_additional_1.map.gz emd_72370_additional_1.map.gz emd_72370_half_map_1.map.gz emd_72370_half_map_1.map.gz emd_72370_half_map_2.map.gz emd_72370_half_map_2.map.gz | 59.6 MB 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-72370 http://ftp.pdbj.org/pub/emdb/structures/EMD-72370 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-72370 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-72370 | HTTPS FTP |
-Related structure data
| Related structure data |  9xzsMC  9xzqC  9xzrC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_72370.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_72370.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full map | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.824 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_72370_msk_1.map emd_72370_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: CryoSPARC-sharpened map
| File | emd_72370_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoSPARC-sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A of the full map
| File | emd_72370_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A of the full map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B of the full map
| File | emd_72370_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B of the full map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Ternary complex of two Trm10 molecules bound to tRNA
| Entire | Name: Ternary complex of two Trm10 molecules bound to tRNA |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex of two Trm10 molecules bound to tRNA
| Supramolecule | Name: Ternary complex of two Trm10 molecules bound to tRNA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Transfer RNA (Gly)
| Macromolecule | Name: Transfer RNA (Gly) / type: rna / ID: 1 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 22.83252 KDa |
| Sequence | String: GCGCAAGAGG UUUAGUGGUA AAAUCCAACG UUGCCAUCGU UGGGCCCCCG GUUCGAUUCC GGGCUUGCGC A GENBANK: GENBANK: CP023996.1 |
-Macromolecule #2: tRNA (guanine(9)-N1)-methyltransferase
| Macromolecule | Name: tRNA (guanine(9)-N1)-methyltransferase / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO / EC number: tRNA (guanine9-N1)-methyltransferase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 34.583582 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSNDEINQNE EKVKRTPPLP PVPEGMSKKQ WKKMCKRQRW EENKAKYNAE RRVKKKRLRH ERSAKIQEYI DRGEEVPQEL IREPRINVN QTDSGIEIIL DCSFDELMND KEIVSLSNQV TRAYSANRRA NHFAEIKVAP FDKRLKQRFE TTLKNTNYEN W NHFKFLPD ...String: MSNDEINQNE EKVKRTPPLP PVPEGMSKKQ WKKMCKRQRW EENKAKYNAE RRVKKKRLRH ERSAKIQEYI DRGEEVPQEL IREPRINVN QTDSGIEIIL DCSFDELMND KEIVSLSNQV TRAYSANRRA NHFAEIKVAP FDKRLKQRFE TTLKNTNYEN W NHFKFLPD DKIMFGDEHI SKDKIVYLTA DTEEKLEKLE PGMRYIVGGI VDKNRYKELC LKKAQKMGIP TRRLPIDEYI NL EGRRVLT TTHVVQLMLK YFDDHNWKNA FESVLPPRKL DAEAKSASSS PAPKDT UniProtKB: tRNA (guanine(9)-N1)-methyltransferase |
-Macromolecule #3: 5'-{[(3S)-3-amino-3-carboxypropyl](ethyl)amino}-5'-deoxyadenosine
| Macromolecule | Name: 5'-{[(3S)-3-amino-3-carboxypropyl](ethyl)amino}-5'-deoxyadenosine type: ligand / ID: 3 / Number of copies: 1 / Formula: AN6 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 395.414 Da |
| Chemical component information | 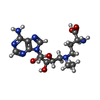 ChemComp-AN6: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: UltrAuFoil / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY ARRAY / Support film - Film thickness: 50 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 11520 pixel / Digitization - Dimensions - Height: 8184 pixel / Number real images: 26645 / Average exposure time: 2.0 sec. / Average electron dose: 59.82 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: Other / Chain - Initial model type: integrative model Details: chain B of the model is from PDB 4JWJ. Chain A was prepared using AlphaFold. |
|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
| Output model |  PDB-9xzs: |
 Movie
Movie Controller
Controller






 Z
Z Y
Y X
X

































