+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7121 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human TRPV6 in amphipols | |||||||||
 Map data Map data | primary map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ion channel / Membrane protein / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationparathyroid hormone secretion / regulation of calcium ion-dependent exocytosis / TRP channels / calcium ion import across plasma membrane / calcium ion homeostasis / calcium channel complex / response to calcium ion / calcium ion transmembrane transport / calcium channel activity / calcium ion transport ...parathyroid hormone secretion / regulation of calcium ion-dependent exocytosis / TRP channels / calcium ion import across plasma membrane / calcium ion homeostasis / calcium channel complex / response to calcium ion / calcium ion transmembrane transport / calcium channel activity / calcium ion transport / calmodulin binding / metal ion binding / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | McGoldrick LL / Singh AK | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2018 Journal: Nature / Year: 2018Title: Opening of the human epithelial calcium channel TRPV6. Authors: Luke L McGoldrick / Appu K Singh / Kei Saotome / Maria V Yelshanskaya / Edward C Twomey / Robert A Grassucci / Alexander I Sobolevsky /  Abstract: Calcium-selective transient receptor potential vanilloid subfamily member 6 (TRPV6) channels play a critical role in calcium uptake in epithelial tissues. Altered TRPV6 expression is associated with ...Calcium-selective transient receptor potential vanilloid subfamily member 6 (TRPV6) channels play a critical role in calcium uptake in epithelial tissues. Altered TRPV6 expression is associated with a variety of human diseases, including cancers. TRPV6 channels are constitutively active and their open probability depends on the lipidic composition of the membrane in which they reside; it increases substantially in the presence of phosphatidylinositol 4,5-bisphosphate. Crystal structures of detergent-solubilized rat TRPV6 in the closed state have previously been solved. Corroborating electrophysiological results, these structures demonstrated that the Ca selectivity of TRPV6 arises from a ring of aspartate side chains in the selectivity filter that binds Ca tightly. However, how TRPV6 channels open and close their pores for ion permeation has remained unclear. Here we present cryo-electron microscopy structures of human TRPV6 in the open and closed states. The channel selectivity filter adopts similar conformations in both states, consistent with its explicit role in ion permeation. The iris-like channel opening is accompanied by an α-to-π-helical transition in the pore-lining transmembrane helix S6 at an alanine hinge just below the selectivity filter. As a result of this transition, the S6 helices bend and rotate, exposing different residues to the ion channel pore in the open and closed states. This gating mechanism, which defines the constitutive activity of TRPV6, is, to our knowledge, unique among tetrameric ion channels and provides structural insights for understanding their diverse roles in physiology and disease. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7121.map.gz emd_7121.map.gz | 5.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7121-v30.xml emd-7121-v30.xml emd-7121.xml emd-7121.xml | 17.5 KB 17.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7121.png emd_7121.png | 101.2 KB | ||
| Filedesc metadata |  emd-7121.cif.gz emd-7121.cif.gz | 6.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7121 http://ftp.pdbj.org/pub/emdb/structures/EMD-7121 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7121 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7121 | HTTPS FTP |
-Related structure data
| Related structure data |  6bo9MC  7120C  7122C  7123C  6bo8C  6boaC  6bobC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7121.map.gz / Format: CCP4 / Size: 32.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7121.map.gz / Format: CCP4 / Size: 32.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | primary map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.98 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
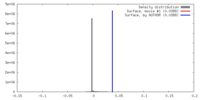
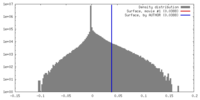
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : TRPV6 ion channel in open states
| Entire | Name: TRPV6 ion channel in open states |
|---|---|
| Components |
|
-Supramolecule #1: TRPV6 ion channel in open states
| Supramolecule | Name: TRPV6 ion channel in open states / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Transient receptor potential cation channel subfamily V member 6
| Macromolecule | Name: Transient receptor potential cation channel subfamily V member 6 type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 85.168953 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGLSLPKEKG LILCLWSKFC RWFQRRESWA QSRDEQNLLQ QKRIWESPLL LAAKDNDVQA LNKLLKYEDC KVHQRGAMGE TALHIAALY DNLEAAMVLM EAAPELVFEP MTSELYEGQT ALHIAVVNQN MNLVRALLAR RASVSARATG TAFRRSPCNL I YFGEHPLS ...String: MGLSLPKEKG LILCLWSKFC RWFQRRESWA QSRDEQNLLQ QKRIWESPLL LAAKDNDVQA LNKLLKYEDC KVHQRGAMGE TALHIAALY DNLEAAMVLM EAAPELVFEP MTSELYEGQT ALHIAVVNQN MNLVRALLAR RASVSARATG TAFRRSPCNL I YFGEHPLS FAACVNSEEI VRLLIEHGAD IRAQDSLGNT VLHILILQPN KTFACQMYNL LLSYDRHGDH LQPLDLVPNH QG LTPFKLA GVEGNTVMFQ HLMQKRKHTQ WTYGPLTSTL YDLTEIDSSG DEQSLLELII TTKKREARQI LDQTPVKELV SLK WKRYGR PYFCMLGAIY LLYIICFTMC CIYRPLKPRT NNRTSPRDNT LLQQKLLQEA YMTPKDDIRL VGELVTVIGA IIIL LVEVP DIFRMGVTRF FGQTILGGPF HVLIITYAFM VLVTMVMRLI SASGEVVPMS FALVLGWCNV MYFARGFQML GPFTI MIQK MIFGDLMRFC WLMAVVILGF ASAFYIIFQT EDPEELGHFY DYPMALFSTF ELFLTIIDGP ANYNVDLPFM YSITYA AFA IIATLLMLNL LIAMMGDTHW RVAHERDELW RAQIVATTVM LERKLPRCLW PRSGICGREY GLGDRWFLRV EDRQDLN RQ RIQRYAQAFH TRGSEDLDKD SVEKLELGCP FSPHLSLPMP SVSRSTSRSS ANWERLRQGT LRRDLRGIIN RGLEDGES W EYQILVPRGS AAAWSHPQFE K UniProtKB: Transient receptor potential cation channel subfamily V member 6 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL |
|---|---|
| Buffer | pH: 8 / Component: (Formula: TRIS, NaCl) / Details: TRIS, NaCl, |
| Grid | Model: C-flat-1.2/1.3 / Material: GOLD / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 67.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER |
|---|---|
| Output model |  PDB-6bo9: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)