+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6915 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SF3b spliceosomal complex bound to E7107 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationU11/U12 snRNP / B-WICH complex / U12-type spliceosomal complex / RNA splicing, via transesterification reactions / splicing factor binding / U2-type precatalytic spliceosome / U2-type prespliceosome assembly / U2-type spliceosomal complex / SAGA complex / U2 snRNP ...U11/U12 snRNP / B-WICH complex / U12-type spliceosomal complex / RNA splicing, via transesterification reactions / splicing factor binding / U2-type precatalytic spliceosome / U2-type prespliceosome assembly / U2-type spliceosomal complex / SAGA complex / U2 snRNP / U2-type prespliceosome / positive regulation of transcription by RNA polymerase III / precatalytic spliceosome / regulation of RNA splicing / spliceosomal complex assembly / positive regulation of transcription by RNA polymerase I / mRNA Splicing - Minor Pathway / U2 snRNA binding / regulation of DNA repair / catalytic step 2 spliceosome / mRNA Splicing - Major Pathway / RNA splicing / stem cell differentiation / spliceosomal complex / mRNA splicing, via spliceosome / negative regulation of protein catabolic process / B-WICH complex positively regulates rRNA expression / nuclear matrix / nuclear speck / chromatin remodeling / mRNA binding / positive regulation of DNA-templated transcription / protein-containing complex binding / nucleolus / positive regulation of transcription by RNA polymerase II / DNA binding / RNA binding / zinc ion binding / nucleoplasm / nucleus Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.95 Å | |||||||||
 Authors Authors | Finci LI / Larsen NA | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Genes Dev / Year: 2018 Journal: Genes Dev / Year: 2018Title: The cryo-EM structure of the SF3b spliceosome complex bound to a splicing modulator reveals a pre-mRNA substrate competitive mechanism of action. Authors: Lorenzo I Finci / Xiaofeng Zhang / Xiuliang Huang / Qiang Zhou / Jennifer Tsai / Teng Teng / Anant Agrawal / Betty Chan / Sean Irwin / Craig Karr / Andrew Cook / Ping Zhu / Dominic Reynolds ...Authors: Lorenzo I Finci / Xiaofeng Zhang / Xiuliang Huang / Qiang Zhou / Jennifer Tsai / Teng Teng / Anant Agrawal / Betty Chan / Sean Irwin / Craig Karr / Andrew Cook / Ping Zhu / Dominic Reynolds / Peter G Smith / Peter Fekkes / Silvia Buonamici / Nicholas A Larsen /   Abstract: Somatic mutations in spliceosome proteins lead to dysregulated RNA splicing and are observed in a variety of cancers. These genetic aberrations may offer a potential intervention point for targeted ...Somatic mutations in spliceosome proteins lead to dysregulated RNA splicing and are observed in a variety of cancers. These genetic aberrations may offer a potential intervention point for targeted therapeutics. SF3B1, part of the U2 small nuclear RNP (snRNP), is targeted by splicing modulators, including E7107, the first to enter clinical trials, and, more recently, H3B-8800. Modulating splicing represents a first-in-class opportunity in drug discovery, and elucidating the structural basis for the mode of action opens up new possibilities for structure-based drug design. Here, we present the cryogenic electron microscopy (cryo-EM) structure of the SF3b subcomplex (SF3B1, SF3B3, PHF5A, and SF3B5) bound to E7107 at 3.95 Å. This structure shows that E7107 binds in the branch point adenosine-binding pocket, forming close contacts with key residues that confer resistance upon mutation: SF3B1 and PHF5A The structure suggests a model in which splicing modulators interfere with branch point adenosine recognition and supports a substrate competitive mechanism of action (MOA). Using several related chemical probes, we validate the pose of the compound and support their substrate competitive MOA by comparing their activity against both strong and weak pre-mRNA substrates. Finally, we present functional data and structure-activity relationship (SAR) on the PHF5A mutation that sensitizes cells to some chemical probes but not others. Developing small molecule splicing modulators represents a promising therapeutic approach for a variety of diseases, and this work provides a significant step in enabling structure-based drug design for these elaborate natural products. Importantly, this work also demonstrates that the utilization of cryo-EM in drug discovery is coming of age. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6915.map.gz emd_6915.map.gz | 28.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6915-v30.xml emd-6915-v30.xml emd-6915.xml emd-6915.xml | 16.7 KB 16.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6915.png emd_6915.png | 152.6 KB | ||
| Masks |  emd_6915_msk_1.map emd_6915_msk_1.map | 30.5 MB |  Mask map Mask map | |
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6915 http://ftp.pdbj.org/pub/emdb/structures/EMD-6915 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6915 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6915 | HTTPS FTP |
-Related structure data
| Related structure data |  5zyaMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6915.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6915.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.322 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_6915_msk_1.map emd_6915_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SF3b Sub-complex
| Entire | Name: SF3b Sub-complex |
|---|---|
| Components |
|
-Supramolecule #1: SF3b Sub-complex
| Supramolecule | Name: SF3b Sub-complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
-Macromolecule #1: Splicing factor 3B subunit 5
| Macromolecule | Name: Splicing factor 3B subunit 5 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 10.149369 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTDRYTIHSQ LEHLQSKYIG TGHADTTKWE WLVNQHRDSY CSYMGHFDLL NYFAIAENES KARVRFNLME KMLQPCGPPA DKPEEN |
-Macromolecule #2: Splicing factor 3B subunit 1
| Macromolecule | Name: Splicing factor 3B subunit 1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 146.024938 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAKIAKTHED IEAQIREIQG KKAALDEAQG VGLDSTGYYD QEIYGGSDSR FAGYVTSIAA TELEDDDDDY SSSTSLLGQK KPGYHAPVA LLNDIPQSTE QYDPFAEHRP PKIADREDEY KKHRRTMIIS PERLDPFADG GKTPDPKMNA RTYMDVMREQ H LTKEEREI ...String: MAKIAKTHED IEAQIREIQG KKAALDEAQG VGLDSTGYYD QEIYGGSDSR FAGYVTSIAA TELEDDDDDY SSSTSLLGQK KPGYHAPVA LLNDIPQSTE QYDPFAEHRP PKIADREDEY KKHRRTMIIS PERLDPFADG GKTPDPKMNA RTYMDVMREQ H LTKEEREI RQQLAEKAKA GELKVVNGAA ASQPPSKRKR RWDQTADQTP GATPKKLSSW DQAETPGHTP SLRWDETPGR AK GSETPGA TPGSKIWDPT PSHTPAGAAT PGRGDTPGHA TPGHGGATSS ARKNRWDETP KTERDTPGHG SGWAETPRTD RGG DSIGET PTPGASKRKS RWDETPASQM GGSTPVLTPG KTPIGTPAMN MATPTPGHIM SMTPEQLQAW RWEREIDERN RPLS DEELD AMFPEGYKVL PPPAGYVPIR TPARKLTATP TPLGGMTGFH MQTEDRTMKS VNDQPSGNLP FLKPDDIQYF DKLLV DVDE STLSPEEQKE RKIMKLLLKI KNGTPPMRKA ALRQITDKAR EFGAGPLFNQ ILPLLMSPTL EDQERHLLVK VIDRIL YKL DDLVRPYVHK ILVVIEPLLI DEDYYARVEG REIISNLAKA AGLATMISTM RPDIDNMDEY VRNTTARAFA VVASALG IP SLLPFLKAVC KSKKSWQARH TGIKIVQQIA ILMGCAILPH LRSLVEIIEH GLVDEQQKVR TISALAIAAL AEAATPYG I ESFDSVLKPL WKGIRQHRGK GLAAFLKAIG YLIPLMDAEY ANYYTREVML ILIREFQSPD EEMKKIVLKV VKQCCGTDG VEANYIKTEI LPPFFKHFWQ HRMALDRRNY RQLVDTTVEL ANKVGAAEII SRIVDDLKDE AEQYRKMVME TIEKIMGNLG AADIDHKLE EQLIDGILYA FQEQTTEDSV MLNGFGTVVN ALGKRVKPYL PQICGTVLWR LNNKSAKVRQ QAADLISRTA V VMKTCQEE KLMGHLGVVL YEYLGEEYPE VLGSILGALK AIVNVIGMHK MTPPIKDLLP RLTPILKNRH EKVQENCIDL VG RIADRGA EYVSAREWMR ICFELLELLK AHKKAIRRAT VNTFGYIAKA IGPHDVLATL LNNLKVQERQ NRVCTTVAIA IVA ETCSPF TVLPALMNEY RVPELNVQNG VLKSLSFLFE YIGEMGKDYI YAVTPLLEDA LMDRDLVHRQ TASAVVQHMS LGVY GFGCE DSLNHLLNYV WPNVFETSPH VIQAVMGALE GLRVAIGPCR MLQYCLQGLF HPARKVRDVY WKIYNSIYIG SQDAL IAHY PRIYNDDKNT YIRYELDYIL |
-Macromolecule #3: PHD finger-like domain-containing protein 5A
| Macromolecule | Name: PHD finger-like domain-containing protein 5A / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 9.394955 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DLIFCRKQAG VAIGRLCEKC DGKCVICDSY VRPCTLVRIC DECNYGSYQG RCVICGGPGV SDAYYCKECT IQEKDRDGCP KIVNL |
-Macromolecule #4: Splicing factor 3B subunit 3
| Macromolecule | Name: Splicing factor 3B subunit 3 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 136.471562 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DMFLYNLTLQ RATGISFAIH GNFSGTKQQE IVVSRGKILE LLRPDPNTGK VHTLLTVEVF GVIRSLMAFR LTGGTKDYIV VGSDSGRIV ILEYQPSKNM FEKIHQETFG KSGCRRIVPG QFLAVDPKGR AVMISAIEKQ KLVYILNRDA AARLTISSPL E AHKANTLV ...String: DMFLYNLTLQ RATGISFAIH GNFSGTKQQE IVVSRGKILE LLRPDPNTGK VHTLLTVEVF GVIRSLMAFR LTGGTKDYIV VGSDSGRIV ILEYQPSKNM FEKIHQETFG KSGCRRIVPG QFLAVDPKGR AVMISAIEKQ KLVYILNRDA AARLTISSPL E AHKANTLV YHVVGVDVGF ENPMFACLEM DYEEADNDPT GEAAANTQQT LTFYELDLGL NHVVRKYSEP LEEHGNFLIT VP GGSDGPS GVLICSENYI TYKNFGDQPD IRCPIPRRRN DLDDPERGMI FVCSATHKTK SMFFFLAQTE QGDIFKITLE TDE DMVTEI RLKYFDTVPV AAAMCVLKTG FLFVASEFGN HYLYQIAHLG DDDEEPEFSS AMPLEEGDTF FFQPRPLKNL VLVD ELDSL SPILFCQIAD LANEDTPQLY VACGRGPRSS LRVLRHGLEV SEMAVSELPG NPNAVWTVRR HIEDEFDAYI IVSFV NATL VLSIGETVEE VTDSGFLGTT PTLSCSLLGD DALVQVYPDG IRHIRADKRV NEWKTPGKKT IVKCAVNQRQ VVIALT GGE LVYFEMDPSG QLNEYTERKE MSADVVCMSL ANVPPGEQRS RFLAVGLVDN TVRIISLDPS DCLQPLSMQA LPAQPES LC IVEMGGTEKQ DELGERGSIG FLYLNIGLQN GVLLRTVLDP VTGDLSDTRT RYLGSRPVKL FRVRMQGQEA VLAMSSRS W LSYSYQSRFH LTPLSYETLE FASGFASEQC PEGIVAISTN TLRILALEKL GAVFNQVAFP LQYTPRKFVI HPESNNLII IETDHNAYTE ATKAQRKQQM AEEMVEAAGE DERELAAEMA AAFLNENLPE SIFGAPKAGN GQWASVIRVM NPIQGNTLDL VQLEQNEAA FSVAVCRFSN TGEDWYVLVG VAKDLILNPR SVAGGFVYTY KLVNNGEKLE FLHKTPVEEV PAAIAPFQGR V LIGVGKLL RVYDLGKKKL LRKCENKHIA NYISGIQTIG HRVIVSDVQE SFIWVRYKRN ENQLIIFADD TYPRWVTTAS LL DYDTVAG ADKFGNICVV RLPPNTNDEV DEDPTGNKAL WDRGLLNGAS QKAEVIMNYH VGETVLSLQK TTLIPGGSES LVY TTLSGG IGILVPFTSH EDHDFFQHVE MHLRSEHPPL CGRDHLSFRS YYFPVKNVID GDLCEQFNSM EPNKQKNVSE ELDR TPPEV SKKLEDIRTR YAFDYKDD |
-Macromolecule #5: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 5 / Number of copies: 3 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #6: [(2~{S},3~{S},4~{E},6~{S},7~{R},10~{R})-3,7-dimethyl-2-[(2~{E},4~...
| Macromolecule | Name: [(2~{S},3~{S},4~{E},6~{S},7~{R},10~{R})-3,7-dimethyl-2-[(2~{E},4~{E},6~{R})-6-methyl-6-oxidanyl-7-[(2~{R},3~{R})-3-[(2~{R},3~{S})-3-oxidanylpentan-2-yl]oxiran-2-yl]hepta-2,4-dien-2-yl]-7,10- ...Name: [(2~{S},3~{S},4~{E},6~{S},7~{R},10~{R})-3,7-dimethyl-2-[(2~{E},4~{E},6~{R})-6-methyl-6-oxidanyl-7-[(2~{R},3~{R})-3-[(2~{R},3~{S})-3-oxidanylpentan-2-yl]oxiran-2-yl]hepta-2,4-dien-2-yl]-7,10-bis(oxidanyl)-12-oxidanylidene-1-oxacyclododec-4-en-6-yl] 4-cycloheptylpiperazine-1-carboxylate type: ligand / ID: 6 / Number of copies: 1 / Formula: 9B0 |
|---|---|
| Molecular weight | Theoretical: 718.96 Da |
| Chemical component information | 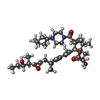 ChemComp-9B0: |
-Macromolecule #7: POTASSIUM ION
| Macromolecule | Name: POTASSIUM ION / type: ligand / ID: 7 / Number of copies: 1 / Formula: K |
|---|---|
| Molecular weight | Theoretical: 39.098 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 55.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.95 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 241288 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)