+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6779 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | GDP-microtubule complexed with nucleotide-free KIF5C | |||||||||||||||
 Map data Map data | KIF5C(nucleotide-free) and Microtubule(GDP) | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Microtubule / KIF5C / Kinesin / STRUCTURAL PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationmotile cilium / structural constituent of cytoskeleton / microtubule cytoskeleton organization / neuron migration / mitotic cell cycle / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / microtubule / hydrolase activity / GTPase activity / GTP binding ...motile cilium / structural constituent of cytoskeleton / microtubule cytoskeleton organization / neuron migration / mitotic cell cycle / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / microtubule / hydrolase activity / GTPase activity / GTP binding / metal ion binding / cytoplasm Similarity search - Function | |||||||||||||||
| Biological species |   | |||||||||||||||
| Method | helical reconstruction / cryo EM / Resolution: 5.35 Å | |||||||||||||||
 Authors Authors | Morikawa M / Shigematsu H / Nitta R | |||||||||||||||
| Funding support |  Japan, 4 items Japan, 4 items
| |||||||||||||||
 Citation Citation |  Journal: J Cell Biol / Year: 2018 Journal: J Cell Biol / Year: 2018Title: Kinesin-binding-triggered conformation switching of microtubules contributes to polarized transport. Authors: Tomohiro Shima / Manatsu Morikawa / Junichi Kaneshiro / Taketoshi Kambara / Shinji Kamimura / Toshiki Yagi / Hiroyuki Iwamoto / Sotaro Uemura / Hideki Shigematsu / Mikako Shirouzu / Taro ...Authors: Tomohiro Shima / Manatsu Morikawa / Junichi Kaneshiro / Taketoshi Kambara / Shinji Kamimura / Toshiki Yagi / Hiroyuki Iwamoto / Sotaro Uemura / Hideki Shigematsu / Mikako Shirouzu / Taro Ichimura / Tomonobu M Watanabe / Ryo Nitta / Yasushi Okada / Nobutaka Hirokawa /   Abstract: Kinesin-1, the founding member of the kinesin superfamily of proteins, is known to use only a subset of microtubules for transport in living cells. This biased use of microtubules is proposed as the ...Kinesin-1, the founding member of the kinesin superfamily of proteins, is known to use only a subset of microtubules for transport in living cells. This biased use of microtubules is proposed as the guidance cue for polarized transport in neurons, but the underlying mechanisms are still poorly understood. Here, we report that kinesin-1 binding changes the microtubule lattice and promotes further kinesin-1 binding. This high-affinity state requires the binding of kinesin-1 in the nucleotide-free state. Microtubules return to the initial low-affinity state by washing out the binding kinesin-1 or by the binding of non-hydrolyzable ATP analogue AMPPNP to kinesin-1. X-ray fiber diffraction, fluorescence speckle microscopy, and second-harmonic generation microscopy, as well as cryo-EM, collectively demonstrated that the binding of nucleotide-free kinesin-1 to GDP microtubules changes the conformation of the GDP microtubule to a conformation resembling the GTP microtubule. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6779.map.gz emd_6779.map.gz | 28.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6779-v30.xml emd-6779-v30.xml emd-6779.xml emd-6779.xml | 15.2 KB 15.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6779.png emd_6779.png | 313 KB | ||
| Filedesc metadata |  emd-6779.cif.gz emd-6779.cif.gz | 6.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6779 http://ftp.pdbj.org/pub/emdb/structures/EMD-6779 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6779 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6779 | HTTPS FTP |
-Related structure data
| Related structure data |  5xxtMC  6781C  6782C  6783C  5xxvC  5xxwC  5xxxC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6779.map.gz / Format: CCP4 / Size: 30.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6779.map.gz / Format: CCP4 / Size: 30.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | KIF5C(nucleotide-free) and Microtubule(GDP) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.284 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
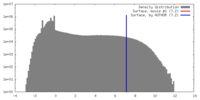
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : GDP-microtubule complexed with nucleotide-free KIF5C
| Entire | Name: GDP-microtubule complexed with nucleotide-free KIF5C |
|---|---|
| Components |
|
-Supramolecule #1: GDP-microtubule complexed with nucleotide-free KIF5C
| Supramolecule | Name: GDP-microtubule complexed with nucleotide-free KIF5C / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Tubulin alpha-1A chain
| Macromolecule | Name: Tubulin alpha-1A chain / type: protein_or_peptide / ID: 1 / Number of copies: 9 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 48.638793 KDa |
| Sequence | String: RECISIHVGQ AGVQIGNACW ELYCLEHGIQ PDGQMPSDKT IGGGDDSFNT FFSETGAGKH VPRAVFVDLE PTVIDEVRTG TYRQLFHPE QLITGKEDAA NNYARGHYTI GKEIIDLVLD RIRKLADQCT GLQGFSVFHS FGGGTGSGFT SLLMERLSVD Y GKKSKLEF ...String: RECISIHVGQ AGVQIGNACW ELYCLEHGIQ PDGQMPSDKT IGGGDDSFNT FFSETGAGKH VPRAVFVDLE PTVIDEVRTG TYRQLFHPE QLITGKEDAA NNYARGHYTI GKEIIDLVLD RIRKLADQCT GLQGFSVFHS FGGGTGSGFT SLLMERLSVD Y GKKSKLEF SIYPAPQVST AVVEPYNSIL TTHTTLEHSD CAFMVDNEAI YDICRRNLDI ERPTYTNLNR LIGQIVSSIT AS LRFDGAL NVDLTEFQTN LVPYPRGHFP LATYAPVISA EKAYHEQLSV AEITNACFEP ANQMVKCDPR HGKYMACCLL YRG DVVPKD VNAAIATIKT KRTIQFVDWC PTGFKVGINY EPPTVVPGGD LAKVQRAVCM LSNTTAIAEA WARLDHKFDL MYAK RAFVH WYVGEGMEEG EFSEAREDMA ALEKDYEEVG VDS UniProtKB: Tubulin alpha-1A chain |
-Macromolecule #2: Tubulin beta chain
| Macromolecule | Name: Tubulin beta chain / type: protein_or_peptide / ID: 2 / Number of copies: 9 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 47.809746 KDa |
| Sequence | String: REIVHIQAGQ CGNQIGAKFW EVISDEHGID PTGSYHGDSD LQLERINVYY NEAAGNKYVP RAILVDLEPG TMDSVRSGPF GQIFRPDNF VFGQSGAGNN WAKGHYTEGA ELVDSVLDVV RKESESCDCL QGFQLTHSLG GGTGSGMGTL LISKIREEYP D RIMNTFSV ...String: REIVHIQAGQ CGNQIGAKFW EVISDEHGID PTGSYHGDSD LQLERINVYY NEAAGNKYVP RAILVDLEPG TMDSVRSGPF GQIFRPDNF VFGQSGAGNN WAKGHYTEGA ELVDSVLDVV RKESESCDCL QGFQLTHSLG GGTGSGMGTL LISKIREEYP D RIMNTFSV VPSPKVSDTV VEPYNATLSV HQLVENTDET YCIDNEALYD ICFRTLKLTT PTYGDLNHLV SATMSGVTTC LR FPGQLNA DLRKLAVNMV PFPRLHFFMP GFAPLTSRGS QQYRALTVPE LTQQMFDAKN MMAACDPRHG RYLTVAAVFR GRM SMKEVD EQMLNVQNKN SSYFVEWIPN NVKTAVCDIP PRGLKMSATF IGNSTAIQEL FKRISEQFTA MFRRKAFLHW YTGE GMDEM EFTEAESNMN DLVSEYQQYQ D UniProtKB: Tubulin beta chain |
-Macromolecule #3: GUANOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 9 / Formula: GTP |
|---|---|
| Molecular weight | Theoretical: 523.18 Da |
| Chemical component information |  ChemComp-GTP: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 9 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: GUANOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-DIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 9 / Formula: GDP |
|---|---|
| Molecular weight | Theoretical: 443.201 Da |
| Chemical component information |  ChemComp-GDP: |
-Macromolecule #6: water
| Macromolecule | Name: water / type: ligand / ID: 6 / Number of copies: 36 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 300 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 8.70662 Å Applied symmetry - Helical parameters - Δ&Phi: -25.7617 ° Applied symmetry - Helical parameters - Axial symmetry: C1 (asymmetric) Resolution.type: BY AUTHOR / Resolution: 5.35 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: FREALIGN (ver. 9) / Number images used: 17173 |
|---|---|
| Startup model | Type of model: EMDB MAP EMDB ID: |
| Final angle assignment | Type: NOT APPLICABLE |
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-5xxt: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)