+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6669 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The endolysosomal Ca2+ channel TRPML1 | |||||||||
 Map data Map data | TrpML1 density map at 8.12A resolution, sharpened by post-processing procedure in RELION | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 8.12 Å | |||||||||
 Authors Authors | Li X / Zhou X | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2017 Journal: Nat Struct Mol Biol / Year: 2017Title: Structural basis of dual Ca/pH regulation of the endolysosomal TRPML1 channel. Authors: Minghui Li / Wei K Zhang / Nicole M Benvin / Xiaoyuan Zhou / Deyuan Su / Huan Li / Shu Wang / Ioannis E Michailidis / Liang Tong / Xueming Li / Jian Yang /   Abstract: The activities of organellar ion channels are often regulated by Ca and H, which are present in high concentrations in many organelles. Here we report a structural element critical for dual Ca/pH ...The activities of organellar ion channels are often regulated by Ca and H, which are present in high concentrations in many organelles. Here we report a structural element critical for dual Ca/pH regulation of TRPML1, a Ca-release channel crucial for endolysosomal function. TRPML1 mutations cause mucolipidosis type IV (MLIV), a severe lysosomal storage disorder characterized by neurodegeneration, mental retardation and blindness. We obtained crystal structures of the 213-residue luminal domain of human TRPML1 containing three missense MLIV-causing mutations. This domain forms a tetramer with a highly electronegative central pore formed by a novel luminal pore loop. Cysteine cross-linking and cryo-EM analyses confirmed that this architecture occurs in the full-length channel. Structure-function studies demonstrated that Ca and H interact with the luminal pore and exert physiologically important regulation. The MLIV-causing mutations disrupt the luminal-domain structure and cause TRPML1 mislocalization. Our study reveals the structural underpinnings of TRPML1's regulation, assembly and pathogenesis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6669.map.gz emd_6669.map.gz | 348.3 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6669-v30.xml emd-6669-v30.xml emd-6669.xml emd-6669.xml | 16 KB 16 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6669.png emd_6669.png | 83.6 KB | ||
| Others |  emd_6669_additional.map.gz emd_6669_additional.map.gz | 981.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6669 http://ftp.pdbj.org/pub/emdb/structures/EMD-6669 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6669 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6669 | HTTPS FTP |
-Validation report
| Summary document |  emd_6669_validation.pdf.gz emd_6669_validation.pdf.gz | 78.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_6669_full_validation.pdf.gz emd_6669_full_validation.pdf.gz | 77.9 KB | Display | |
| Data in XML |  emd_6669_validation.xml.gz emd_6669_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6669 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6669 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6669 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6669 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6669.map.gz / Format: CCP4 / Size: 2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6669.map.gz / Format: CCP4 / Size: 2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | TrpML1 density map at 8.12A resolution, sharpened by post-processing procedure in RELION | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.64 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: TrpML1 density map, not sharpened
| File | emd_6669_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | TrpML1 density map, not sharpened | ||||||||||||
| Projections & Slices |
| ||||||||||||
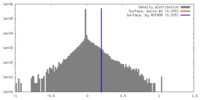
| Density Histograms |
- Sample components
Sample components
-Entire : The endolysosomal Ca2+ channel TRPML1
| Entire | Name: The endolysosomal Ca2+ channel TRPML1 |
|---|---|
| Components |
|
-Supramolecule #1: The endolysosomal Ca2+ channel TRPML1
| Supramolecule | Name: The endolysosomal Ca2+ channel TRPML1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) / Recombinant plasmid: pFastBac1 Trichoplusia ni (cabbage looper) / Recombinant plasmid: pFastBac1 |
-Macromolecule #1: The endolysosomal Ca2+ channel TRPML1
| Macromolecule | Name: The endolysosomal Ca2+ channel TRPML1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GGGGSMSRRS TTDRSDFNDN ASNASSNASR RPTINFQEIM EDLPHHERET GERLRRHLQF FFMNPMEKWK VRHQLPYKLV LQVLKIVFVT MQLILFAEMR MSHVDFLEDT TTVMRHRFLK EWNDDRDALQ YPPAEGRYSV YDDQGLSEHL SFLINSYYSI RNDSFASFSY ...String: GGGGSMSRRS TTDRSDFNDN ASNASSNASR RPTINFQEIM EDLPHHERET GERLRRHLQF FFMNPMEKWK VRHQLPYKLV LQVLKIVFVT MQLILFAEMR MSHVDFLEDT TTVMRHRFLK EWNDDRDALQ YPPAEGRYSV YDDQGLSEHL SFLINSYYSI RNDSFASFSY DVVSHPSGNL GAQISFESIP PIEVLIDRIS NVTVNNNTYN FDIREVKDTK RLNLTETEVF QIGQSDDAVR DILATRGITF LPEDALKIST VQFKFRLRTI HYSPTAGDQK PECYKISVSI KFDNSRHTGQ VHVTLSTVVS YVNVCNGRII KGVGWSFDTL LIGGTDIFVL ILCILSLILC CRALIKAHLL QIKTSDYFEN VLKNKITVTD QLDFLNLWYV MIVVNDALII IGTVAKISIE FQDFDNSLFT LTSIFLGMGA LLVYVGVLRY FGFFSQYNIL MLTLKRSAPN IMRFMTCAIV LYAGFLIAGW VIIGPYSMKF RTLAESSEAL FSLLNGDDMF ATFYTINDSN TVIKVFGTVY IYLFVSLFIY VVLSLFIAII MDAYEVVKDR YSDGLRAIEK RGCLRDFVES NPPPSELGSP TTRSAYAPSN LLNLGRGWQR LE |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.6 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||
| Grid | Model: R1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.039 kPa | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281.15 K / Instrument: FEI VITROBOT MARK IV Details: waiting for 3 seconds before blotting for 3.5 seconds(double-sided,blot force 1), then the grid was immediately plunged into liquid ethane cooled by liquid-nitrogen.. | |||||||||
| Details | the sample was homogeneous |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 7676 pixel / Digitization - Dimensions - Height: 7420 pixel / Digitization - Sampling interval: 5.0 µm / Digitization - Frames/image: 1-32 / Average exposure time: 8.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.1 µm / Nominal defocus min: 2.1 µm / Nominal magnification: 22500 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)