[English] 日本語
 Yorodumi
Yorodumi- EMDB-66120: Cryo-EM structure of Candida glabrata GPI mannosyltransferase I b... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Candida glabrata GPI mannosyltransferase I bound to Dol-P-Man | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GT-C / GPI / Mannosyltransferase / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationdol-P-Man:GlcN-acyl-PI alpha-1,4-mannosyltransferase activity / glycosylphosphatidylinositol-mannosyltransferase I complex / mannosyltransferase activity / GPI anchor biosynthetic process / fungal-type cell wall organization / Transferases; Glycosyltransferases; Hexosyltransferases / ERAD pathway / protein processing / endoplasmic reticulum membrane Similarity search - Function | |||||||||
| Biological species |  Nakaseomyces glabratus (fungus) Nakaseomyces glabratus (fungus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.48 Å | |||||||||
 Authors Authors | Sun H / Wu WH / Yan ZF | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: J Fungi (Basel) / Year: 2025 Journal: J Fungi (Basel) / Year: 2025Title: Structural Insights into the Glycosylphosphatidylinositol Mannosyltransferase I Complex from . Authors: Hui Sun / Weihong Wu / Xiaomei Li / Yang Deng / Jiarong Huang / Meng Yin / Zhaofeng Yan /  Abstract: The global rise in resistance to first-line antifungal agents highlights the urgent need for new therapeutic strategies. Glycosylphosphatidylinositol (GPI)-anchored protein biosynthesis is an ...The global rise in resistance to first-line antifungal agents highlights the urgent need for new therapeutic strategies. Glycosylphosphatidylinositol (GPI)-anchored protein biosynthesis is an attractive target. The GPI mannosyltransferase I (GPI-MT-I), composed of Gpi14 and Pbn1, catalyzes the essential first mannose transfer from dolichol-phosphomannose (Dol-P-Man) to the GPI precursor. This initial mannosylation is critical for fungal cell wall integrity, yet the molecular basis of GPI-MT-I assembly and substrate recognition remains poorly understood. Here, we present the cryo-EM structure of GPI-MT-I in complex with Dol-P-Man, revealing how Gpi14 and Pbn1 form a stable complex and engage the mannose donor. An AlphaFold3-predicted acceptor-bound model further defines the structural basis of acceptor substrate recognition and suggests a plausible catalytic mechanism. Comparison with structural homologs highlights a distinct mode of substrate engagement by GPI-MT-I. Together, these findings establish a mechanistic framework for GPI-MT-I function with broader implications for the GPI-MT family. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_66120.map.gz emd_66120.map.gz | 38.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-66120-v30.xml emd-66120-v30.xml emd-66120.xml emd-66120.xml | 17.8 KB 17.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_66120_fsc.xml emd_66120_fsc.xml | 7.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_66120.png emd_66120.png | 84.7 KB | ||
| Masks |  emd_66120_msk_1.map emd_66120_msk_1.map | 40.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-66120.cif.gz emd-66120.cif.gz | 6.3 KB | ||
| Others |  emd_66120_half_map_1.map.gz emd_66120_half_map_1.map.gz emd_66120_half_map_2.map.gz emd_66120_half_map_2.map.gz | 37.7 MB 37.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-66120 http://ftp.pdbj.org/pub/emdb/structures/EMD-66120 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-66120 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-66120 | HTTPS FTP |
-Related structure data
| Related structure data |  9wnoMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_66120.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_66120.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.088 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_66120_msk_1.map emd_66120_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_66120_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_66120_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : GPI mannosyltransferase I
| Entire | Name: GPI mannosyltransferase I |
|---|---|
| Components |
|
-Supramolecule #1: GPI mannosyltransferase I
| Supramolecule | Name: GPI mannosyltransferase I / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Nakaseomyces glabratus (fungus) Nakaseomyces glabratus (fungus) |
-Macromolecule #1: GPI mannosyltransferase 1
| Macromolecule | Name: GPI mannosyltransferase 1 / type: protein_or_peptide / ID: 1 Details: Sequence reference for strain 'Nakaseomyces glabratus' is not available in UniProt at the time of biocuration. Current sequence reference is from UniProt id Q6FXQ5. Number of copies: 1 / Enantiomer: LEVO EC number: Transferases; Glycosyltransferases; Hexosyltransferases |
|---|---|
| Source (natural) | Organism:  Nakaseomyces glabratus (fungus) Nakaseomyces glabratus (fungus) |
| Molecular weight | Theoretical: 50.289254 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MFKLGTIGLL SLAVLLRIAF FLFGVYQDEH FTVKYTDIDY QVFNDAAWFV AHRKSPYNRD TYRYTPLLSW LLLPNHMIPW FHFGKLIFV LCDLATGVLI LQMLKKLKSK YRYGTDRMTI MAAIWLLNPM VITISTRGNA ESVLCFLILS FLYCFLCEQY L LGGLLFGL ...String: MFKLGTIGLL SLAVLLRIAF FLFGVYQDEH FTVKYTDIDY QVFNDAAWFV AHRKSPYNRD TYRYTPLLSW LLLPNHMIPW FHFGKLIFV LCDLATGVLI LQMLKKLKSK YRYGTDRMTI MAAIWLLNPM VITISTRGNA ESVLCFLILS FLYCFLCEQY L LGGLLFGL SIHFKIYPII YALPIAIYVA AAHYNKTQSV FKSSFKLFLV GFSTLIVLIL LTVFMYMLYG DKFIDQTYLY HI YRTDHRH NFSVWNMLLY FNSALPKTSE LSKFVFLPQM IIVLAISLTQ LRRPSSFPLL CNVLFLETFA FVTFNKVCTS QYF IWYLIF LPFVLYNTRI SMRRGIVMLV VWVATQALWL RQGYLLEFEG QNVFYPGLFC ASVGFFVSNA WILGQFIEDT IIQN DYVYR IEPSESRSKD SAVIDKPAIA KTEKAE UniProtKB: GPI mannosyltransferase 1 |
-Macromolecule #2: Protein PBN1
| Macromolecule | Name: Protein PBN1 / type: protein_or_peptide / ID: 2 Details: Sequence reference for strain 'Nakaseomyces glabratus' is not available in UniProt at the time of biocuration. Current sequence reference is from UniProt id Q6FX62. Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Nakaseomyces glabratus (fungus) Nakaseomyces glabratus (fungus) |
| Molecular weight | Theoretical: 47.799812 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MESVRHRIAL LFANEADIQD VETVEDGVIV FPNSNITIQD RWTYAIDYEL DSIRRISWRN PSSTRQFSVI ESRLAPGFNI YSNDKEARL NLFGIQPVYS PMYKSLHSET WKSINEILPG GKNLNIPWNP ELCDYDILIT ANTVQVFSYC SLVEKKKFVK D AGKVEIGL ...String: MESVRHRIAL LFANEADIQD VETVEDGVIV FPNSNITIQD RWTYAIDYEL DSIRRISWRN PSSTRQFSVI ESRLAPGFNI YSNDKEARL NLFGIQPVYS PMYKSLHSET WKSINEILPG GKNLNIPWNP ELCDYDILIT ANTVQVFSYC SLVEKKKFVK D AGKVEIGL FHVDTEDEED INLSGLRCTW EDTSNNIGKC EKTTLFYKPF HLYVDDSSDI APITIENTNG LHPKMKIDLS GI RKDKDCR HFVFSQLPSE IFVDKFQSPG SIVFGLDDLE LPDYKVNSLS WGSESIVTLK EGQINEITYF SRYLEPKERA SNK TVGFEP ILFKACQVGK EEDLSNPFYS RSLGYEAYFS ENTKFYHINS TSVHVNIPYP NKNDIGNIEY TTFGIVVLAI CYLI YKLLR PSRKLSTVKR D UniProtKB: Protein PBN1 |
-Macromolecule #4: dolichyl phosphate mannose
| Macromolecule | Name: dolichyl phosphate mannose / type: ligand / ID: 4 / Number of copies: 1 / Formula: MJC |
|---|---|
| Molecular weight | Theoretical: 1.011439 KDa |
| Chemical component information | 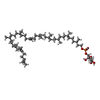 ChemComp-MJC: |
-Macromolecule #5: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 5 / Number of copies: 1 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 1 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 56.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.1 µm / Nominal defocus min: 1.7 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)













































