+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of rat TRPV1 in complex with SB-366791 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TRPV1 / protein complex / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of establishment of blood-brain barrier / response to capsazepine / sensory perception of mechanical stimulus / cellular response to temperature stimulus / peptide secretion / smooth muscle contraction involved in micturition / excitatory extracellular ligand-gated monoatomic ion channel activity / temperature-gated ion channel activity / detection of chemical stimulus involved in sensory perception of pain / TRP channels ...negative regulation of establishment of blood-brain barrier / response to capsazepine / sensory perception of mechanical stimulus / cellular response to temperature stimulus / peptide secretion / smooth muscle contraction involved in micturition / excitatory extracellular ligand-gated monoatomic ion channel activity / temperature-gated ion channel activity / detection of chemical stimulus involved in sensory perception of pain / TRP channels / fever generation / detection of temperature stimulus involved in thermoception / urinary bladder smooth muscle contraction / thermoception / cellular response to acidic pH / negative regulation of systemic arterial blood pressure / response to pH / chloride channel regulator activity / dendritic spine membrane / monoatomic cation transmembrane transporter activity / glutamate secretion / ligand-gated monoatomic ion channel activity / negative regulation of heart rate / response to pain / temperature homeostasis / diet induced thermogenesis / cellular response to alkaloid / cellular response to ATP / cellular response to cytokine stimulus / detection of temperature stimulus involved in sensory perception of pain / intracellularly gated calcium channel activity / behavioral response to pain / negative regulation of mitochondrial membrane potential / calcium ion import across plasma membrane / monoatomic cation channel activity / extracellular ligand-gated monoatomic ion channel activity / sensory perception of pain / phosphatidylinositol binding / phosphoprotein binding / microglial cell activation / cellular response to nerve growth factor stimulus / response to peptide hormone / calcium ion transmembrane transport / GABA-ergic synapse / lipid metabolic process / cellular response to growth factor stimulus / calcium channel activity / positive regulation of nitric oxide biosynthetic process / cellular response to tumor necrosis factor / transmembrane signaling receptor activity / calcium ion transport / sensory perception of taste / cellular response to heat / positive regulation of cytosolic calcium ion concentration / response to heat / monoatomic ion transmembrane transport / protein homotetramerization / postsynaptic membrane / calmodulin binding / neuron projection / positive regulation of apoptotic process / external side of plasma membrane / neuronal cell body / dendrite / negative regulation of transcription by RNA polymerase II / ATP binding / metal ion binding / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.25 Å | |||||||||
 Authors Authors | Chen X / Yu Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM structure of rat TRPV1 in complex with the analgesic drug SB-36679 Authors: Yu Y | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_65639.map.gz emd_65639.map.gz | 59.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-65639-v30.xml emd-65639-v30.xml emd-65639.xml emd-65639.xml | 15.6 KB 15.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_65639_fsc.xml emd_65639_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_65639.png emd_65639.png | 119.8 KB | ||
| Filedesc metadata |  emd-65639.cif.gz emd-65639.cif.gz | 5.9 KB | ||
| Others |  emd_65639_half_map_1.map.gz emd_65639_half_map_1.map.gz emd_65639_half_map_2.map.gz emd_65639_half_map_2.map.gz | 59.1 MB 59.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-65639 http://ftp.pdbj.org/pub/emdb/structures/EMD-65639 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-65639 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-65639 | HTTPS FTP |
-Related structure data
| Related structure data |  9w4nMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_65639.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_65639.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.93 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_65639_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_65639_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of rat TRPV1 in complex with the analgesic drug...
| Entire | Name: Cryo-EM structure of rat TRPV1 in complex with the analgesic drug SB-36679 |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of rat TRPV1 in complex with the analgesic drug...
| Supramolecule | Name: Cryo-EM structure of rat TRPV1 in complex with the analgesic drug SB-36679 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Transient receptor potential cation channel subfamily V member 1
| Macromolecule | Name: Transient receptor potential cation channel subfamily V member 1 type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 99.514625 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DYKDDDDKWS HPQFEKGGGG SGGSAWSHPQ FEKEFKGLVD MEQRASLDSE ESESPPQENS CLDPPDRDPN CKPPPVKPHI FTTRSRTRL FGKGDSEEAS PLDCPYEEGG LASCPIITVS SVLTIQRPGD GPASVRPSSQ DSVSAGEKPP RLYDRRSIFD A VAQSNCQE ...String: DYKDDDDKWS HPQFEKGGGG SGGSAWSHPQ FEKEFKGLVD MEQRASLDSE ESESPPQENS CLDPPDRDPN CKPPPVKPHI FTTRSRTRL FGKGDSEEAS PLDCPYEEGG LASCPIITVS SVLTIQRPGD GPASVRPSSQ DSVSAGEKPP RLYDRRSIFD A VAQSNCQE LESLLPFLQR SKKRLTDSEF KDPETGKTCL LKAMLNLHNG QNDTIALLLD VARKTDSLKQ FVNASYTDSY YK GQTALHI AIERRNMTLV TLLVENGADV QAAANGDFFK KTKGRPGFYF GELPLSLAAC TNQLAIVKFL LQNSWQPADI SAR DSVGNT VLHALVEVAD NTVDNTKFVT SMYNEILILG AKLHPTLKLE EITNRKGLTP LALAASSGKI GVLAYILQRE IHEP ECRHL SRKFTEWAYG PVHSSLYDLS CIDTCEKNSV LEVIAYSSSE TPNRHDMLLV EPLNRLLQDK WDRFVKRIFY FNFFV YCLY MIIFTAAAYY RPVEGLPPYK LKNTVGDYFR VTGEILSVSG GVYFFFRGIQ YFLQRRPSLK SLFVDSYSEI LFFVQS LFM LVSVVLYFSQ RKEYVASMVF SLAMGWTNML YYTRGFQQMG IYAVMIEKMI LRDLCRFMFV YLVFLFGFST AVVTLIE DG KNNSLPMEST PHKCRGSACK PGNSYNSLYS TCLELFKFTI GMGDLEFTEN YDFKAVFIIL LLAYVILTYI LLLNMLIA L MGETVNKIAQ ESKNIWKLQR AITILDTEKS FLKCMRKAFR SGKLLQVGFT PDGKDDYRWC FRVDEVNWTT WNTNVGIIN EDPGNCEGVK RTLSFSLRSG RVSGRNWKNF ALVPLLRDAS TRDRHATQQE EVQLKHYTGS LKPEDAEVFK DSMVPGEK UniProtKB: Transient receptor potential cation channel subfamily V member 1 |
-Macromolecule #2: (2E)-3-(4-chlorophenyl)-N-(3-methoxyphenyl)prop-2-enamide
| Macromolecule | Name: (2E)-3-(4-chlorophenyl)-N-(3-methoxyphenyl)prop-2-enamide type: ligand / ID: 2 / Number of copies: 4 / Formula: ZEI |
|---|---|
| Molecular weight | Theoretical: 287.741 Da |
| Chemical component information | 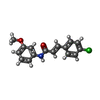 ChemComp-ZEI: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 / Details: 20 mM Hepes, 150 mM NaCl |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average exposure time: 4.5 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





































