+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The TUG-891-bound structure of TMEM175 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TMEM175 / cro-EM / TUG-891 / Parkinson / lysosome / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationlysosomal lumen pH elevation / phagosome-lysosome fusion / regulation of lysosomal lumen pH / potassium ion leak channel activity / proton channel activity / arachidonate binding / potassium channel activity / potassium ion transmembrane transport / proton transmembrane transport / neuron cellular homeostasis ...lysosomal lumen pH elevation / phagosome-lysosome fusion / regulation of lysosomal lumen pH / potassium ion leak channel activity / proton channel activity / arachidonate binding / potassium channel activity / potassium ion transmembrane transport / proton transmembrane transport / neuron cellular homeostasis / lysosome / endosome / endosome membrane / lysosomal membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.74 Å | |||||||||
 Authors Authors | Zhu X / Liu H / Yin W | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Neuron / Year: 2025 Journal: Neuron / Year: 2025Title: Structural insights into the activation of TMEM175 by small molecule. Authors: Xuewu Zhu / Meixuan Ping / Heng Liu / Ting Yu / Zhongwen Jiang / Zhenhua Liu / Chanjing Li / Xinjiao Hou / Qinyu Chu / Shuyao Li / Caiwen Mao / Ting Luo / Chunlan Kang / Feng Wang / Chuanyan ...Authors: Xuewu Zhu / Meixuan Ping / Heng Liu / Ting Yu / Zhongwen Jiang / Zhenhua Liu / Chanjing Li / Xinjiao Hou / Qinyu Chu / Shuyao Li / Caiwen Mao / Ting Luo / Chunlan Kang / Feng Wang / Chuanyan Yang / Meiqin Tang / Zhidong Jiang / Zhaobing Gao / Hong Liu / H Eric Xu / Beisha Tang / Xi Cheng / Wanchao Yin / Yu Zhou / Ping Li /  Abstract: The upregulation of transmembrane protein 175 (TMEM175) has the potential to improve Parkinson's disease (PD) by aiding in the removal of α-synuclein aggregates. Understanding the structural basis ...The upregulation of transmembrane protein 175 (TMEM175) has the potential to improve Parkinson's disease (PD) by aiding in the removal of α-synuclein aggregates. Understanding the structural basis of TMEM175 agonisms is crucial for uncovering its therapeutic potential for PD. Here, we have identified the first cryo-electron microscopy (cryo-EM) structure of human TMEM175 complexes with three agonists: DCY1020, DCY1040, and TUG-891. An open state of TMEM175 is unequivocally captured, laying the groundwork for designing more effective agonists. Further investigations using surface plasmon resonance, systematic mutagenesis, whole-endolysosome patch-clamp techniques, and molecular dynamics simulations consistently revealed that DCY1020/1040 binds at the interface between two subunits, inducing an open conformation further augmented by the synergistic agonist TUG-891. Notably, these agonists facilitate the removal of pathological α-synuclein and restore functions of PD-related TMEM175 variants in neurons. Our findings provide proof of concept that drug discovery targeting TMEM175 can develop agonists capable of effectively reducing pathological α-synuclein levels in PD. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_65304.map.gz emd_65304.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-65304-v30.xml emd-65304-v30.xml emd-65304.xml emd-65304.xml | 17.6 KB 17.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_65304.png emd_65304.png | 74.8 KB | ||
| Masks |  emd_65304_msk_1.map emd_65304_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-65304.cif.gz emd-65304.cif.gz | 6 KB | ||
| Others |  emd_65304_half_map_1.map.gz emd_65304_half_map_1.map.gz emd_65304_half_map_2.map.gz emd_65304_half_map_2.map.gz | 59.2 MB 59.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-65304 http://ftp.pdbj.org/pub/emdb/structures/EMD-65304 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-65304 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-65304 | HTTPS FTP |
-Related structure data
| Related structure data |  9vsqMC  9vspC  9vsrC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_65304.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_65304.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.824 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_65304_msk_1.map emd_65304_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_65304_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_65304_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : DCY1020-bound structure of TMEM175
| Entire | Name: DCY1020-bound structure of TMEM175 |
|---|---|
| Components |
|
-Supramolecule #1: DCY1020-bound structure of TMEM175
| Supramolecule | Name: DCY1020-bound structure of TMEM175 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Endosomal/lysosomal proton channel TMEM175
| Macromolecule | Name: Endosomal/lysosomal proton channel TMEM175 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 55.667219 KDa |
| Recombinant expression | Organism: Mammalian expression vector EGFP-MCS-pcDNA3.1 (others) |
| Sequence | String: MSQPRTPEQA LDTPGDCPPG RRDEDAGEGI QCSQRMLSFS DALLSIIATV MILPVTHTEI SPEQQFDRSV QRLLATRIAV YLMTFLIVT VAWAAHTRLF QVVGKTDDTL ALLNLACMMT ITFLPYTFSL MVTFPDVPLG IFLFCVCVIA IGVVQALIVG Y AFHFPHLL ...String: MSQPRTPEQA LDTPGDCPPG RRDEDAGEGI QCSQRMLSFS DALLSIIATV MILPVTHTEI SPEQQFDRSV QRLLATRIAV YLMTFLIVT VAWAAHTRLF QVVGKTDDTL ALLNLACMMT ITFLPYTFSL MVTFPDVPLG IFLFCVCVIA IGVVQALIVG Y AFHFPHLL SPQIQRSAHR ALYRRHVLGI VLQGPALCFA AAIFSLFFVP LSYLLMVTVI LLPYVSKVTG WCRDRLLGHR EP SAHPVEV FSFDLHEPLS KERVEAFSDG VYAIVATLLI LDICEDNVPD PKDVKERFSG SLVAALSATG PRFLAYFGSF ATV GLLWFA HHSLFLHVRK ATRAMGLLNT LSLAFVGGLP LAYQQTSAFA RQPRDELERV RVSCTIIFLA SIFQLAMWTT ALLH QAETL QPSVWFGGRE HVLMFAKLAL YPCASLLAFA STCLLSRFSV GIFHLMQIAV PCAFLLLRLL VGLALATLRV LRGLA RPEH PPPAPTGQDD PQSQLLPAPC UniProtKB: Endosomal/lysosomal proton channel TMEM175 |
-Macromolecule #2: 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}prop...
| Macromolecule | Name: 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid type: ligand / ID: 2 / Number of copies: 2 / Formula: YN9 |
|---|---|
| Molecular weight | Theoretical: 364.409 Da |
| Chemical component information | 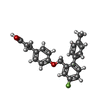 ChemComp-YN9: |
-Macromolecule #3: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 3 / Number of copies: 2 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Macromolecule #4: POTASSIUM ION
| Macromolecule | Name: POTASSIUM ION / type: ligand / ID: 4 / Number of copies: 7 / Formula: K |
|---|---|
| Molecular weight | Theoretical: 39.098 Da |
-Macromolecule #5: water
| Macromolecule | Name: water / type: ligand / ID: 5 / Number of copies: 40 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 1.38 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DARK FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)













































