+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Helical structure of KomBC in complex with dITP | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | KomBc / dITP / Sir2 / CELL INVASION | |||||||||
| Function / homology |  Function and homology information Function and homology informationnucleoside triphosphate catabolic process / nucleoside triphosphate diphosphatase activity / nucleotide metabolic process / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Archangium gephyra (bacteria) Archangium gephyra (bacteria) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 3.04 Å | |||||||||
 Authors Authors | Li Y / Zheng Q / Li S | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2026 Journal: Mol Cell / Year: 2026Title: Filament-driven activation of the Kongming antiviral system by deoxyinosine triphosphate. Authors: Xiangkai Zhen / Yu Li / Zihe Liu / Yang Huang / Xurong Wang / Shuying Xu / Yuchen Jiang / Fan Li / Jinfu Su / Qi Lai / Shaowei Li / Ningshao Xia / Qingbing Zheng / Songying Ouyang /  Abstract: Nucleotide-derived second messengers are frequently deployed by bacteria to activate effector proteins to mediate the immunity. The Kongming system uses deoxyinosine triphosphate (dITP) to trigger ...Nucleotide-derived second messengers are frequently deployed by bacteria to activate effector proteins to mediate the immunity. The Kongming system uses deoxyinosine triphosphate (dITP) to trigger nicotinamide adenine dinucleotide (NAD) depletion via the Sir2-domain protein KomC. We reveal that dITP binding to the KomB-KomC (KomBC) complex stabilizes KomB dimerization, initiating hierarchical allosteric changes. This drives KomBC filament assembly, which is essential for activating the NADase activity of KomC. Cryo-EM structures of apo-, dITP-bound, NAD-bound and postcatalytic KomBC filaments show the structural landscape of how dITP-induced remodeling reshapes the catalytic pocket of KomC, enabling NAD hydrolysis. Mutagenesis confirms that filament assembly and allostery are critical for catalysis. These findings elucidate the structural basis for the recognition of the nucleotide derivative signaling molecule, the assembly and the filament-mediated allosteric activation mechanism in prokaryotic immunity and a distinct variation of Sir2 NADase activation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_64788.map.gz emd_64788.map.gz | 169.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-64788-v30.xml emd-64788-v30.xml emd-64788.xml emd-64788.xml | 23.8 KB 23.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_64788.png emd_64788.png | 102.6 KB | ||
| Filedesc metadata |  emd-64788.cif.gz emd-64788.cif.gz | 6.1 KB | ||
| Others |  emd_64788_additional_1.map.gz emd_64788_additional_1.map.gz emd_64788_additional_2.map.gz emd_64788_additional_2.map.gz emd_64788_half_map_1.map.gz emd_64788_half_map_1.map.gz emd_64788_half_map_2.map.gz emd_64788_half_map_2.map.gz | 168.7 MB 165.7 MB 318.5 MB 318.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-64788 http://ftp.pdbj.org/pub/emdb/structures/EMD-64788 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-64788 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-64788 | HTTPS FTP |
-Related structure data
| Related structure data |  9v57MC  9v0gC  9v0yC  9v0zC  9v12C  9v2kC  9v2mC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_64788.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_64788.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.778 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_64788_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #2
| File | emd_64788_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_64788_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_64788_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : KomBC filament with dITP
| Entire | Name: KomBC filament with dITP |
|---|---|
| Components |
|
-Supramolecule #1: KomBC filament with dITP
| Supramolecule | Name: KomBC filament with dITP / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Archangium gephyra (bacteria) Archangium gephyra (bacteria) |
-Macromolecule #1: Xanthosine/inosine triphosphate pyrophosphatase
| Macromolecule | Name: Xanthosine/inosine triphosphate pyrophosphatase / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Archangium gephyra (bacteria) Archangium gephyra (bacteria) |
| Molecular weight | Theoretical: 20.012025 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAPTWFYNTT NSEKLRELQH VLGGSAKLGY LTAKVTEILD VDLETVIRAK AIAAYRAVRV PVIVEHGALC IDALNGLPGA LVKPFWESL DTRLCEVIPA GQRTARARGA LCYCDGRERH VLIEETEGEI APSARGTGGF HWDPIFIPKG QTRTFAEMSL D EKLSFSPL GRLHTRLRTE LGL UniProtKB: XTP/dITP diphosphohydrolase |
-Macromolecule #2: NAD-dependent protein deacetylase
| Macromolecule | Name: NAD-dependent protein deacetylase / type: protein_or_peptide / ID: 2 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Archangium gephyra (bacteria) Archangium gephyra (bacteria) |
| Molecular weight | Theoretical: 31.370715 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTTLTLSEAA PLLKKEFREG RLIPFLGAGF SKPLKLPDGS QLIASLAKTL GFEPELFDMH GRFEQLAEFF AISAPNRLQR LVYEMSLSF DSAEAEALRE KSPMHRALAA LDWRTIYTTN YDKHVEGALR DAGKQAAVLA SFADFQGPRA RDVCEVIKFH G TLDQPDTI ...String: MTTLTLSEAA PLLKKEFREG RLIPFLGAGF SKPLKLPDGS QLIASLAKTL GFEPELFDMH GRFEQLAEFF AISAPNRLQR LVYEMSLSF DSAEAEALRE KSPMHRALAA LDWRTIYTTN YDKHVEGALR DAGKQAAVLA SFADFQGPRA RDVCEVIKFH G TLDQPDTI VLTESSYFQR MALDAPPDQR LRADLLANSF LFIGYSFSDT NIRYIWYRMN QLREQSQLGV KHSQARRCFF AT HGAGLVQ PDILQQWNID VIQLDPTDKS ASVARLLESI A UniProtKB: NAD-dependent protein deacetylase |
-Macromolecule #3: 2'-deoxyinosine 5'-triphosphate
| Macromolecule | Name: 2'-deoxyinosine 5'-triphosphate / type: ligand / ID: 3 / Number of copies: 8 / Formula: Y43 |
|---|---|
| Molecular weight | Theoretical: 492.166 Da |
| Chemical component information | 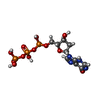 ChemComp-Y43: |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 8 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 49.25 Å Applied symmetry - Helical parameters - Δ&Phi: 83.37 ° Applied symmetry - Helical parameters - Axial symmetry: C2 (2 fold cyclic) Resolution.type: BY AUTHOR / Resolution: 3.04 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 1574792 |
|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION |
| Startup model | Type of model: OTHER |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




















































