[English] 日本語
 Yorodumi
Yorodumi- EMDB-6430: Characterization of red-shifted phycobiliprotein complexes isolat... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6430 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Characterization of red-shifted phycobiliprotein complexes isolated from the chlorophyll f-containing cyanobacterium Halomicronema hongdechloris | |||||||||
 Map data Map data | Reconstruction of red-shifted phycobiliprotein complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | phycobiliprotein / complementary chromatic acclimation / far-red light / small angle neutron scattering / photosynthesis / cyanobacteria | |||||||||
| Function / homology |  Function and homology information Function and homology informationphycobilisome / plasma membrane-derived thylakoid membrane / photosynthesis Similarity search - Function | |||||||||
| Biological species |  Halomicronema hongdechloris (bacteria) Halomicronema hongdechloris (bacteria) | |||||||||
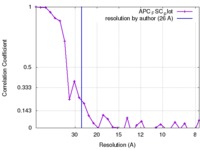
| Method | single particle reconstruction / negative staining / Resolution: 26.0 Å | |||||||||
 Authors Authors | Li Y / Lin Y / Garvey C / Birch D / Corkery RW / Loughlin PC / Scheer H / Willows RD / Chen M | |||||||||
 Citation Citation |  Journal: Biochim Biophys Acta / Year: 2016 Journal: Biochim Biophys Acta / Year: 2016Title: Characterization of red-shifted phycobilisomes isolated from the chlorophyll f-containing cyanobacterium Halomicronema hongdechloris. Authors: Yaqiong Li / Yuankui Lin / Christopher J Garvey / Debra Birch / Robert W Corkery / Patrick C Loughlin / Hugo Scheer / Robert D Willows / Min Chen /    Abstract: Phycobilisomes are the main light-harvesting protein complexes in cyanobacteria and some algae. It is commonly accepted that these complexes only absorb green and orange light, complementing ...Phycobilisomes are the main light-harvesting protein complexes in cyanobacteria and some algae. It is commonly accepted that these complexes only absorb green and orange light, complementing chlorophyll absorbance. Here, we present a new phycobilisome derived complex that consists only of allophycocyanin core subunits, having red-shifted absorption peaks of 653 and 712 nm. These red-shifted phycobiliprotein complexes were isolated from the chlorophyll f-containing cyanobacterium, Halomicronema hongdechloris, grown under monochromatic 730 nm-wavelength (far-red) light. The 3D model obtained from single particle analysis reveals a double disk assembly of 120-145 Å with two α/β allophycocyanin trimers fitting into the two separated disks. They are significantly smaller than typical phycobilisomes formed from allophycocyanin subunits and core-membrane linker proteins, which fit well with a reduced distance between thylakoid membranes observed from cells grown under far-red light. Spectral analysis of the dissociated and denatured phycobiliprotein complexes grown under both these light conditions shows that the same bilin chromophore, phycocyanobilin, is exclusively used. Our findings show that red-shifted phycobilisomes are required for assisting efficient far-red light harvesting. Their discovery provides new insights into the molecular mechanisms of light harvesting under extreme conditions for photosynthesis, as well as the strategies involved in flexible chromatic acclimation to diverse light conditions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6430.map.gz emd_6430.map.gz | 680.4 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6430-v30.xml emd-6430-v30.xml emd-6430.xml emd-6430.xml | 12.3 KB 12.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_6430_fsc.xml emd_6430_fsc.xml | 2.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_6430.jpg emd_6430.jpg | 10.9 KB | ||
| Masks |  emd_6430_msk_1.map emd_6430_msk_1.map | 844.8 KB |  Mask map Mask map | |
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6430 http://ftp.pdbj.org/pub/emdb/structures/EMD-6430 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6430 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6430 | HTTPS FTP |
-Related structure data
| Related structure data |  3jbbMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6430.map.gz / Format: CCP4 / Size: 1.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6430.map.gz / Format: CCP4 / Size: 1.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of red-shifted phycobiliprotein complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.8 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Segmentation: Mask volume for reconstruction
| Annotation | Mask volume for reconstruction | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| File |  emd_6430_msk_1.map emd_6430_msk_1.map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Red-shifted phycobilisomes from Halomicronema hongdechloris
| Entire | Name: Red-shifted phycobilisomes from Halomicronema hongdechloris |
|---|---|
| Components |
|
-Supramolecule #1000: Red-shifted phycobilisomes from Halomicronema hongdechloris
| Supramolecule | Name: Red-shifted phycobilisomes from Halomicronema hongdechloris type: sample / ID: 1000 / Oligomeric state: Two APC trimers linked with APC E / Number unique components: 1 |
|---|
-Macromolecule #1: APC phycobilisome complex
| Macromolecule | Name: APC phycobilisome complex / type: protein_or_peptide / ID: 1 / Details: Taxonomy ID 1209493 / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  Halomicronema hongdechloris (bacteria) Halomicronema hongdechloris (bacteria) |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: 0.8 M phosphate buffer |
|---|---|
| Staining | Type: NEGATIVE / Details: 2% uranyl acetate for 2-3 seconds |
| Grid | Details: 200 mesh gold grid with thin carbon support |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | OTHER |
|---|---|
| Date | Jun 8, 2015 |
| Image recording | Category: CCD / Film or detector model: GENERIC IMAGE PLATES / Number real images: 12 |
| Electron beam | Acceleration voltage: 100 kV / Electron source: TUNGSTEN HAIRPIN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal magnification: 180000 |
| Sample stage | Specimen holder model: PHILIPS ROTATION HOLDER |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - #0 - Chain ID: A / Chain - #1 - Chain ID: B |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | Two trimers were fitted to each end of the EM map. 98% of atoms fit within the map at contour level 0.87. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-3jbb: |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)