[English] 日本語
 Yorodumi
Yorodumi- EMDB-6412: Structure of PhnGI complex from Escherichia coli by negative stain -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6412 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of PhnGI complex from Escherichia coli by negative stain | |||||||||
 Map data Map data | PhnGI complex from Escherichia coli | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | PhnGI | |||||||||
| Biological species |  | |||||||||
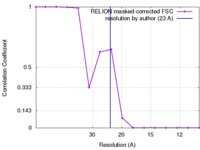
| Method | single particle reconstruction / negative staining / Resolution: 23.0 Å | |||||||||
 Authors Authors | Yang K / Ren Z / Raushel FM / Zhang J | |||||||||
 Citation Citation |  Journal: Structure / Year: 2016 Journal: Structure / Year: 2016Title: Structures of the Carbon-Phosphorus Lyase Complex Reveal the Binding Mode of the NBD-like PhnK. Authors: Kailu Yang / Zhongjie Ren / Frank M Raushel / Junjie Zhang /  Abstract: The carbon-phosphorus (C-P) lyase complex is essential for the metabolism of unactivated phosphonates to phosphate in bacteria. Using single-particle cryo-electron microscopy, we determined two ...The carbon-phosphorus (C-P) lyase complex is essential for the metabolism of unactivated phosphonates to phosphate in bacteria. Using single-particle cryo-electron microscopy, we determined two structures of the C-P lyase core complex PhnG2H2I2J2, with or without PhnK. PhnG2H2I2J2 is a two-fold symmetric hetero-octamer. Its two PhnJ subunits provide two identical binding sites for PhnK. Only one PhnK binds to PhnG2H2I2J2 due to steric hindrance. PhnK is homologous to the nucleotide-binding domain (NBD) of ATP-binding cassette transporters. The α helices 3 and 4 of PhnK bind to α helix 6 and a loop (residues 227-230) of PhnJ, in a different mode from the binding of NBDs to their transmembrane partners. Moreover, binding of PhnK exposes the active site residue, Gly32 of PhnJ, located near the interface between PhnJ and PhnH. This structural information provides a basis for further deciphering of the reaction mechanism of the C-P lyase. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6412.map.gz emd_6412.map.gz | 30.7 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6412-v30.xml emd-6412-v30.xml emd-6412.xml emd-6412.xml | 9.7 KB 9.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_6412_fsc.xml emd_6412_fsc.xml | 1.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_6412.png emd_6412.png | 25.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6412 http://ftp.pdbj.org/pub/emdb/structures/EMD-6412 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6412 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6412 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6412.map.gz / Format: CCP4 / Size: 104.5 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6412.map.gz / Format: CCP4 / Size: 104.5 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | PhnGI complex from Escherichia coli | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 5.3 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : PhnGI complex from Escherichia coli
| Entire | Name: PhnGI complex from Escherichia coli |
|---|---|
| Components |
|
-Supramolecule #1000: PhnGI complex from Escherichia coli
| Supramolecule | Name: PhnGI complex from Escherichia coli / type: sample / ID: 1000 / Oligomeric state: Two PhnG, two PhnI / Number unique components: 2 |
|---|---|
| Molecular weight | Theoretical: 115 KDa |
-Macromolecule #1: PhnG
| Macromolecule | Name: PhnG / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 18.7 KDa |
| Recombinant expression | Organism:  |
-Macromolecule #2: PhnI
| Macromolecule | Name: PhnI / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 38.9 KDa |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.01 mg/mL |
|---|---|
| Buffer | pH: 8.5 / Details: 50 mM HEPES, 150 mM NaCl, 2 mM TCEP |
| Staining | Type: NEGATIVE / Details: sample was stained by 2% uranyl acetate |
| Grid | Details: 200 mesh copper grid with carbon film |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Date | May 21, 2014 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 1000 (2k x 2k) / Number real images: 20 / Average electron dose: 40 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 26285 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 4.1 µm / Nominal defocus min: 3.0 µm / Nominal magnification: 40000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)