[English] 日本語
 Yorodumi
Yorodumi- EMDB-64036: Cryo-EM structure of the Lhcp trimer from Ostreococcus tauri at 1... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the Lhcp trimer from Ostreococcus tauri at 1.94 angstrom resolution | ||||||||||||||||||||||||
 Map data Map data | |||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords | LHCp / Prasinophytes / green light absorption / carotenoid / photosynthesis / Light-harvesting complex / prasinoxanthin / micromonal / dihydrolutein / uriolide / acyl-antheraxanthin / DVP. | ||||||||||||||||||||||||
| Function / homology | photosynthesis, light harvesting / Chlorophyll A-B binding protein, plant and chromista / Chlorophyll A-B binding protein / Chlorophyll A-B binding protein / photosystem I / photosystem II / chlorophyll binding / chloroplast thylakoid membrane / Chlorophyll a-b binding protein, chloroplastic Function and homology information Function and homology information | ||||||||||||||||||||||||
| Biological species |  Ostreococcus tauri (plant) Ostreococcus tauri (plant) | ||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 1.94 Å | ||||||||||||||||||||||||
 Authors Authors | Seki S / Kubota M / Ishii A / Kim E / Tanaka H / Miyata T / Namba K / Kurisu G / Minagawa J / Fujii R | ||||||||||||||||||||||||
| Funding support |  Japan, 7 items Japan, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM structure of the LHCp trimer from Ostreococcus tauri at 1.94 angstrom resolution Authors: Soichiro S / Kubota M / Ishii A / Kim E / Tomoko M / Hideaki T / Keiichi N / Genji K / Minagawa J / Ritsuko F | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_64036.map.gz emd_64036.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-64036-v30.xml emd-64036-v30.xml emd-64036.xml emd-64036.xml | 26 KB 26 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_64036_fsc.xml emd_64036_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_64036.png emd_64036.png | 60 KB | ||
| Filedesc metadata |  emd-64036.cif.gz emd-64036.cif.gz | 7.5 KB | ||
| Others |  emd_64036_half_map_1.map.gz emd_64036_half_map_1.map.gz emd_64036_half_map_2.map.gz emd_64036_half_map_2.map.gz | 59.5 MB 59.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-64036 http://ftp.pdbj.org/pub/emdb/structures/EMD-64036 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-64036 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-64036 | HTTPS FTP |
-Related structure data
| Related structure data |  9uc6MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_64036.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_64036.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.859 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_64036_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_64036_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Lhcp trimer from Ostreococcus tauri
+Supramolecule #1: Lhcp trimer from Ostreococcus tauri
+Macromolecule #1: Chlorophyll a-b binding protein, chloroplastic
+Macromolecule #2: CHLOROPHYLL A
+Macromolecule #3: CHLOROPHYLL B
+Macromolecule #4: Mg-2,4-divinyl-phaeoporphyrin a5-monometyl ester
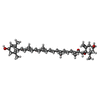
+Macromolecule #5: (3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-1-[(1~{S},4...
+Macromolecule #6: (5~{Z})-3-[2-[(1~{R},4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2...
+Macromolecule #7: (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY...
+Macromolecule #8: (2~{Z},4~{E},6~{E},8~{E},10~{E},12~{E},14~{E},16~{E})-6,11,15-tri...
+Macromolecule #9: (1~{R})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},...
+Macromolecule #10: Lauryl Maltose Neopentyl Glycol
+Macromolecule #11: 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL
+Macromolecule #12: [(1~{S},3~{S},6~{R})-1,5,5-trimethyl-6-[(1~{E},3~{E},5~{E},7~{E},...
+Macromolecule #13: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5.0 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Component - Concentration: 25.0 mM / Component - Formula: C8H18N2O4S / Component - Name: HEPES / Details: 0.003% LMNG |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 10 sec. / Pretreatment - Pressure: 10000.0 kPa |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Specialist optics | Energy filter - Name: In-column Omega Filter / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 6144 / Average exposure time: 3.0 sec. / Average electron dose: 80.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 60000 |
| Sample stage | Specimen holder model: JEOL CRYOSPECPORTER / Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)