[English] 日本語
 Yorodumi
Yorodumi- EMDB-6392: Structural basis for TBC-DEG assembly with soluble tubulin dimer -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6392 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structural basis for TBC-DEG assembly with soluble tubulin dimer | |||||||||
 Map data Map data | Reconstruction of TBC-DEG with tubulin dimer | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Tubulin cofactors / Microtubule dynamics / Chaperones / tubulin biogenesis or degradation / tubulin dimer / TBC-DEG | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 24.0 Å | |||||||||
 Authors Authors | Nithianantham S / Le S / Seto E / Jia W / Leary J / Corbett KD / Moore JK / Al-Bassam J | |||||||||
 Citation Citation |  Journal: Elife / Year: 2015 Journal: Elife / Year: 2015Title: Tubulin cofactors and Arl2 are cage-like chaperones that regulate the soluble αβ-tubulin pool for microtubule dynamics. Authors: Stanley Nithianantham / Sinh Le / Elbert Seto / Weitao Jia / Julie Leary / Kevin D Corbett / Jeffrey K Moore / Jawdat Al-Bassam /  Abstract: Microtubule dynamics and polarity stem from the polymerization of αβ-tubulin heterodimers. Five conserved tubulin cofactors/chaperones and the Arl2 GTPase regulate α- and β-tubulin assembly into ...Microtubule dynamics and polarity stem from the polymerization of αβ-tubulin heterodimers. Five conserved tubulin cofactors/chaperones and the Arl2 GTPase regulate α- and β-tubulin assembly into heterodimers and maintain the soluble tubulin pool in the cytoplasm, but their physical mechanisms are unknown. Here, we reconstitute a core tubulin chaperone consisting of tubulin cofactors TBCD, TBCE, and Arl2, and reveal a cage-like structure for regulating αβ-tubulin. Biochemical assays and electron microscopy structures of multiple intermediates show the sequential binding of αβ-tubulin dimer followed by tubulin cofactor TBCC onto this chaperone, forming a ternary complex in which Arl2 GTP hydrolysis is activated to alter αβ-tubulin conformation. A GTP-state locked Arl2 mutant inhibits ternary complex dissociation in vitro and causes severe defects in microtubule dynamics in vivo. Our studies suggest a revised paradigm for tubulin cofactors and Arl2 functions as a catalytic chaperone that regulates soluble αβ-tubulin assembly and maintenance to support microtubule dynamics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6392.map.gz emd_6392.map.gz | 7.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6392-v30.xml emd-6392-v30.xml emd-6392.xml emd-6392.xml | 12.5 KB 12.5 KB | Display Display |  EMDB header EMDB header |
| Images |  400_6392.gif 400_6392.gif 80_6392.gif 80_6392.gif | 39.2 KB 3.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6392 http://ftp.pdbj.org/pub/emdb/structures/EMD-6392 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6392 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6392 | HTTPS FTP |
-Validation report
| Summary document |  emd_6392_validation.pdf.gz emd_6392_validation.pdf.gz | 78.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_6392_full_validation.pdf.gz emd_6392_full_validation.pdf.gz | 77.5 KB | Display | |
| Data in XML |  emd_6392_validation.xml.gz emd_6392_validation.xml.gz | 495 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6392 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6392 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6392 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6392 | HTTPS FTP |
-Related structure data
| Related structure data |  6390C  6391C  6393C  5cyaC C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10035 (Title: Tubulin Chaperone complexes TBC-DEG Q73L: alpha beta-tubulin complex EMPIAR-10035 (Title: Tubulin Chaperone complexes TBC-DEG Q73L: alpha beta-tubulin complexData size: 1.1 Data #1: CTF-corrected particle images of tubulin chaperone complexes TBC-DEG Q73L: alpha beta-tubulin complex [picked particles - multiframe - processed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6392.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6392.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of TBC-DEG with tubulin dimer | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
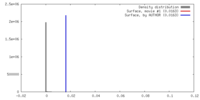
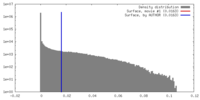
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Tubulin cofactor complex [TBC-DE(N-GFP)G]
| Entire | Name: Tubulin cofactor complex [TBC-DE(N-GFP)G] |
|---|---|
| Components |
|
-Supramolecule #1000: Tubulin cofactor complex [TBC-DE(N-GFP)G]
| Supramolecule | Name: Tubulin cofactor complex [TBC-DE(N-GFP)G] / type: sample / ID: 1000 Details: TBC-DEG complex with tubulin dimer behaves as a single biochemical entity. Oligomeric state: One heterotrimer of TBC-DEG binds to tubulin dimer Number unique components: 5 |
|---|---|
| Molecular weight | Experimental: 308 KDa / Theoretical: 310 KDa / Method: SEC-MALS |
-Macromolecule #1: tubulin folding cofactor D
| Macromolecule | Name: tubulin folding cofactor D / type: protein_or_peptide / ID: 1 / Name.synonym: TBCD / Number of copies: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Macromolecule #2: tubulin folding cofactor E
| Macromolecule | Name: tubulin folding cofactor E / type: protein_or_peptide / ID: 2 / Name.synonym: TBCE / Number of copies: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Macromolecule #3: ARL2 GTPase, Q73L mutant
| Macromolecule | Name: ARL2 GTPase, Q73L mutant / type: protein_or_peptide / ID: 3 / Name.synonym: DEG(Q73L) / Details: Arl2 locked mutant (Q73L) in a GTP-like state / Number of copies: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Macromolecule #4: alpha tubulin
| Macromolecule | Name: alpha tubulin / type: protein_or_peptide / ID: 4 / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism: unidentified (others) |
-Macromolecule #5: beta tubulin
| Macromolecule | Name: beta tubulin / type: protein_or_peptide / ID: 5 / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism: unidentified (others) |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7 Details: 50 mM HEPES, 100 mM KCl, 0.1 mM GTP, 3 mM b-mercaptoethanol |
| Staining | Type: NEGATIVE Details: The proteins were incubated on carbon-coated grids, briefly washed, and stained with 1% uranyl formate. |
| Grid | Details: glow-discharged 200 mesh gold grid with thin carbon support |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2100F |
|---|---|
| Date | Jul 10, 2014 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: PRIMESCAN / Digitization - Sampling interval: 3.5 µm / Number real images: 90 / Average electron dose: 9 e/Å2 / Bits/pixel: 5 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 50050 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.7 µm / Nominal defocus min: 1.1 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder model: JEOL |
- Image processing
Image processing
| Details | TBC-DEG:tubulin complex particles were picked semi-automatically using e2boxer.py. |
|---|---|
| CTF correction | Details: Each particle |
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 24.0 Å / Resolution method: OTHER / Software - Name: Spider, EMAN2 / Number images used: 20000 |
| Final two d classification | Number classes: 400 |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)